Bar plot
Bior_BarPlot.RdCreate a stacked barplot.
Bior_BarPlot(
data,
x,
y,
combine = FALSE,
merge = FALSE,
color = "black",
fill = "white",
palette = NULL,
size = NULL,
width = NULL,
title = NULL,
xlab = NULL,
ylab = NULL,
facet.by = NULL,
panel.labs = NULL,
short.panel.labs = TRUE,
select = NULL,
remove = NULL,
order = NULL,
add = "none",
add.params = list(),
error.plot = "errorbar",
label = FALSE,
lab.col = "black",
lab.size = 4,
lab.pos = c("out", "in"),
lab.vjust = NULL,
lab.hjust = NULL,
lab.nb.digits = NULL,
sort.val = c("none", "desc", "asc"),
sort.by.groups = TRUE,
top = Inf,
position = position_stack(),
ggtheme = theme_pubr(),
...
)Arguments
- data
a data frame
- x, y
x and y variables for drawing.
- combine
logical value. Default is FALSE. Used only when y is a vector containing multiple variables to plot. If TRUE, create a multi-panel plot by combining the plot of y variables.
- merge
logical or character value. Default is FALSE. Used only when y is a vector containing multiple variables to plot. If TRUE, merge multiple y variables in the same plotting area. Allowed values include also "asis" (TRUE) and "flip". If merge = "flip", then y variables are used as x tick labels and the x variable is used as grouping variable.
- color, fill
outline and fill colors.
- palette
the color palette to be used for coloring or filling by groups. Allowed values include "grey" for grey color palettes; brewer palettes e.g. "RdBu", "Blues", ...; or custom color palette e.g. c("blue", "red"); and scientific journal palettes from ggsci R package, e.g.: "npg", "aaas", "lancet", "jco", "ucscgb", "uchicago", "simpsons" and "rickandmorty".
- size
Numeric value (e.g.: size = 1). change the size of points and outlines.
- width
numeric value between 0 and 1 specifying box width.
- title
plot main title.
- xlab
character vector specifying x axis labels. Use xlab = FALSE to hide xlab.
- ylab
character vector specifying y axis labels. Use ylab = FALSE to hide ylab.
- facet.by
character vector, of length 1 or 2, specifying grouping variables for faceting the plot into multiple panels. Should be in the data.
- panel.labs
a list of one or two character vectors to modify facet panel labels. For example, panel.labs = list(sex = c("Male", "Female")) specifies the labels for the "sex" variable. For two grouping variables, you can use for example panel.labs = list(sex = c("Male", "Female"), rx = c("Obs", "Lev", "Lev2") ).
- short.panel.labs
logical value. Default is TRUE. If TRUE, create short labels for panels by omitting variable names; in other words panels will be labelled only by variable grouping levels.
- select
character vector specifying which items to display.
- remove
character vector specifying which items to remove from the plot.
- order
character vector specifying the order of items.
- add
character vector for adding another plot element (e.g.: dot plot or error bars). Allowed values are one or the combination of: "none", "dotplot", "jitter", "boxplot", "point", "mean", "mean_se", "mean_sd", "mean_ci", "mean_range", "median", "median_iqr", "median_hilow", "median_q1q3", "median_mad", "median_range"; see ?desc_statby for more details.
- add.params
parameters (color, shape, size, fill, linetype) for the argument 'add'; e.g.: add.params = list(color = "red").
- error.plot
plot type used to visualize error. Allowed values are one of c("pointrange", "linerange", "crossbar", "errorbar", "upper_errorbar", "lower_errorbar", "upper_pointrange", "lower_pointrange", "upper_linerange", "lower_linerange"). Default value is "pointrange" or "errorbar". Used only when add != "none" and add contains one "mean_*" or "med_*" where "*" = sd, se, ....
- label
specify whether to add labels on the bar plot. Allowed values are:
logical value: If TRUE, y values is added as labels on the bar plot
character vector: Used as text labels; must be the same length as y.
- lab.col, lab.size
text color and size for labels.
- lab.pos
character specifying the position for labels. Allowed values are "out" (for outside) or "in" (for inside). Ignored when lab.vjust != NULL.
- lab.vjust
numeric, vertical justification of labels. Provide negative value (e.g.: -0.4) to put labels outside the bars or positive value to put labels inside (e.g.: 2).
- lab.hjust
numeric, horizontal justification of labels.
- lab.nb.digits
integer indicating the number of decimal places (round) to be used.
- sort.val
a string specifying whether the value should be sorted. Allowed values are "none" (no sorting), "asc" (for ascending) or "desc" (for descending).
- sort.by.groups
logical value. If TRUE the data are sorted by groups. Used only when sort.val != "none".
- top
a numeric value specifying the number of top elements to be shown.
- position
Position adjustment, either as a string naming the adjustment (e.g.
"jitter"to useposition_jitter), or the result of a call to a position adjustment function. Use the latter if you need to change the settings of the adjustment.- ggtheme
function, ggplot2 theme name. Default value is theme_pubr(). Allowed values include ggplot2 official themes: theme_gray(), theme_bw(), theme_minimal(), theme_classic(), theme_void(), ....
- ...
other arguments to be passed to be passed to ggpar().
Value
A ggplot object
Examples
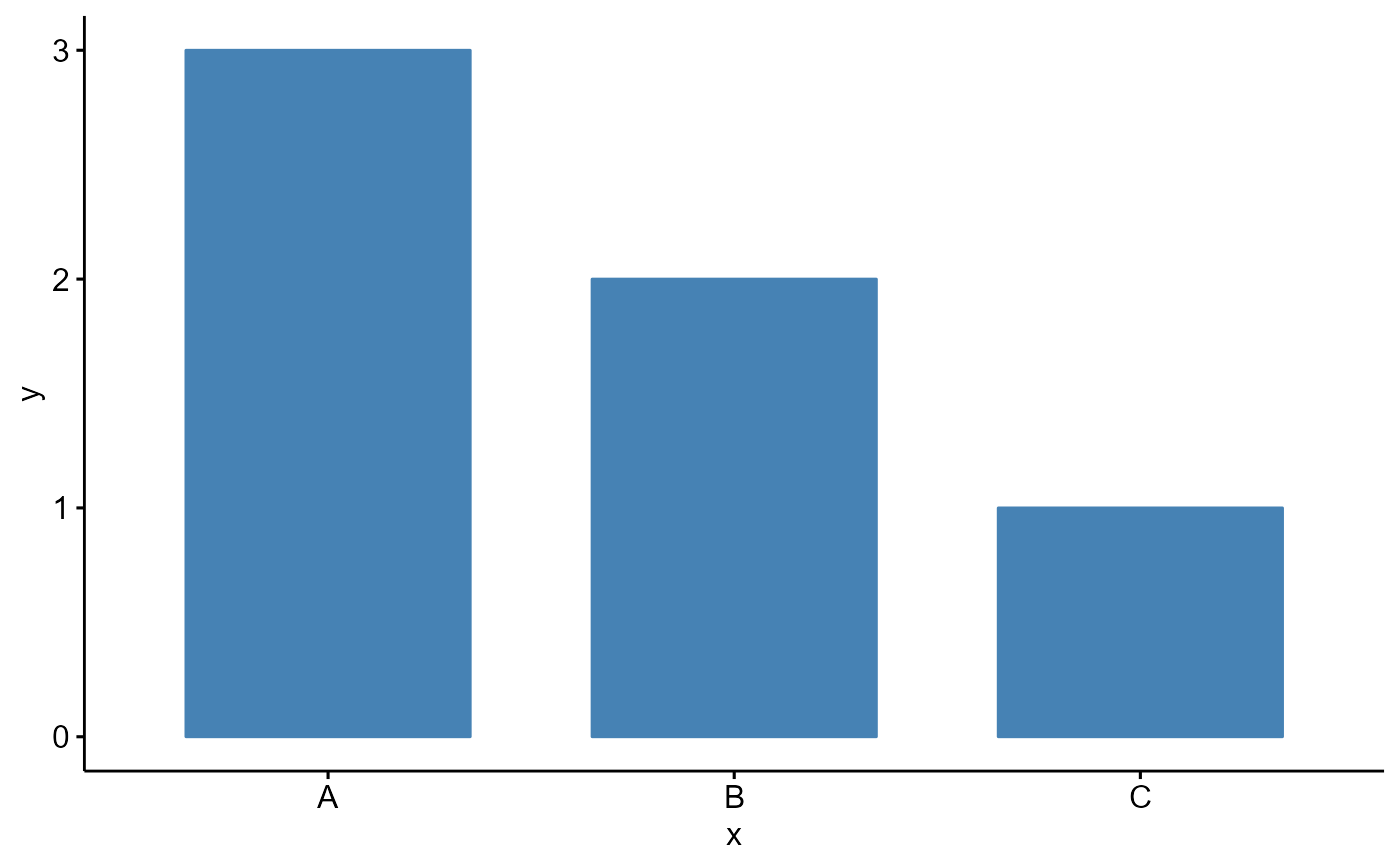
# Examples 1
df <- data.frame(
x = c("A", "B", "C"),
y = c(3, 2, 1))
Bior_BarPlot(df, "x", "y", fill = "steelblue", color = "steelblue")
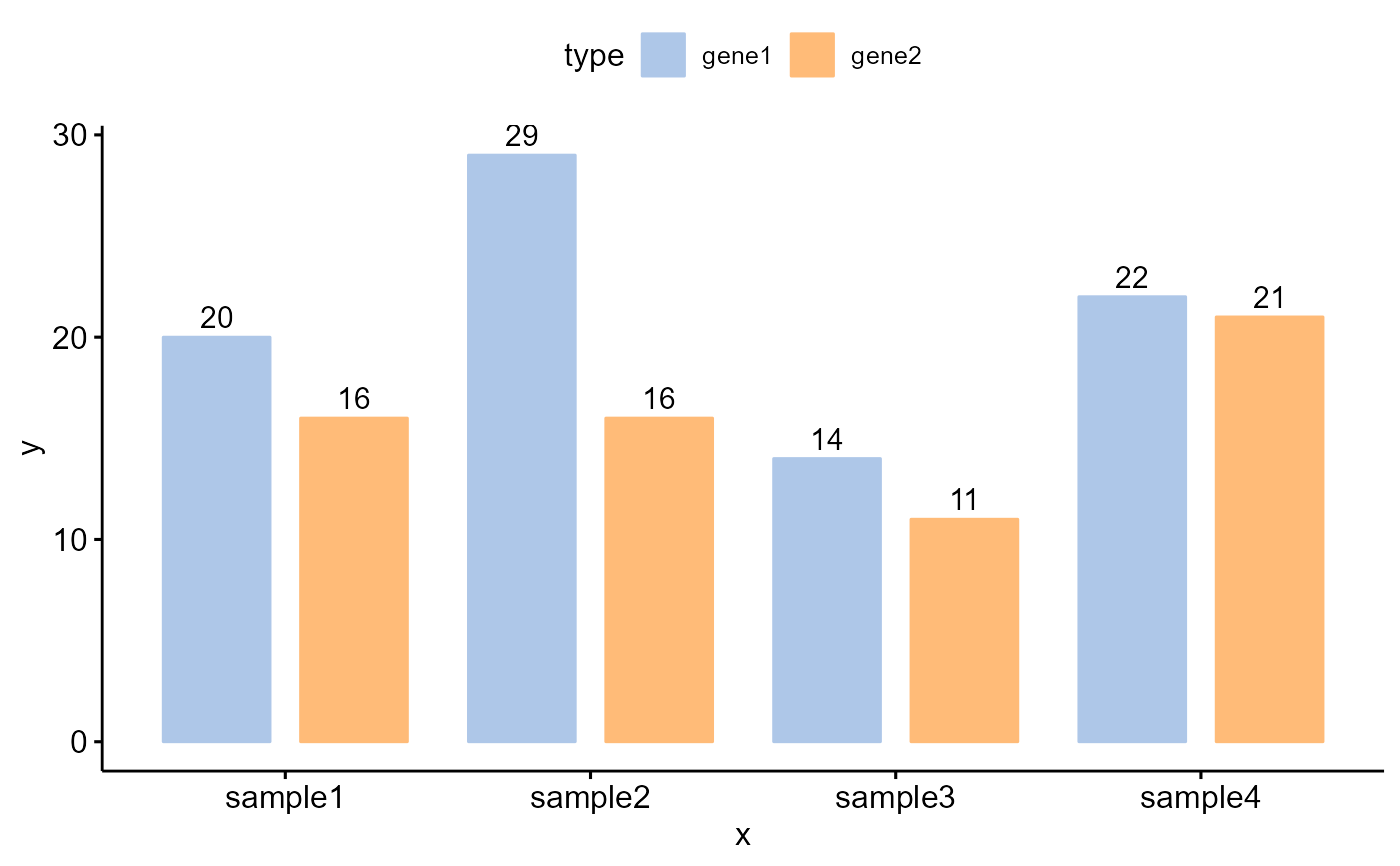
 # Examples 2
df <- data.frame(
x = rep(c('sample1','sample2','sample3','sample4'), each=2),
y = c(20,16,29,16,14,11,22,21),
type = rep(c('gene1','gene2'), 4),
label = c(20,16,29,16,14,11,"","")
)
col <- c("#AEC7E8FF","#FFBB78FF")
Bior_BarPlot(df, "x", "y", fill = "type", color = "type", label = df$label,
palette = col, lab.pos = "in") +
theme(legend.position = "right", legend.key.size=unit(1, "cm"))
# Examples 2
df <- data.frame(
x = rep(c('sample1','sample2','sample3','sample4'), each=2),
y = c(20,16,29,16,14,11,22,21),
type = rep(c('gene1','gene2'), 4),
label = c(20,16,29,16,14,11,"","")
)
col <- c("#AEC7E8FF","#FFBB78FF")
Bior_BarPlot(df, "x", "y", fill = "type", color = "type", label = df$label,
palette = col, lab.pos = "in") +
theme(legend.position = "right", legend.key.size=unit(1, "cm"))
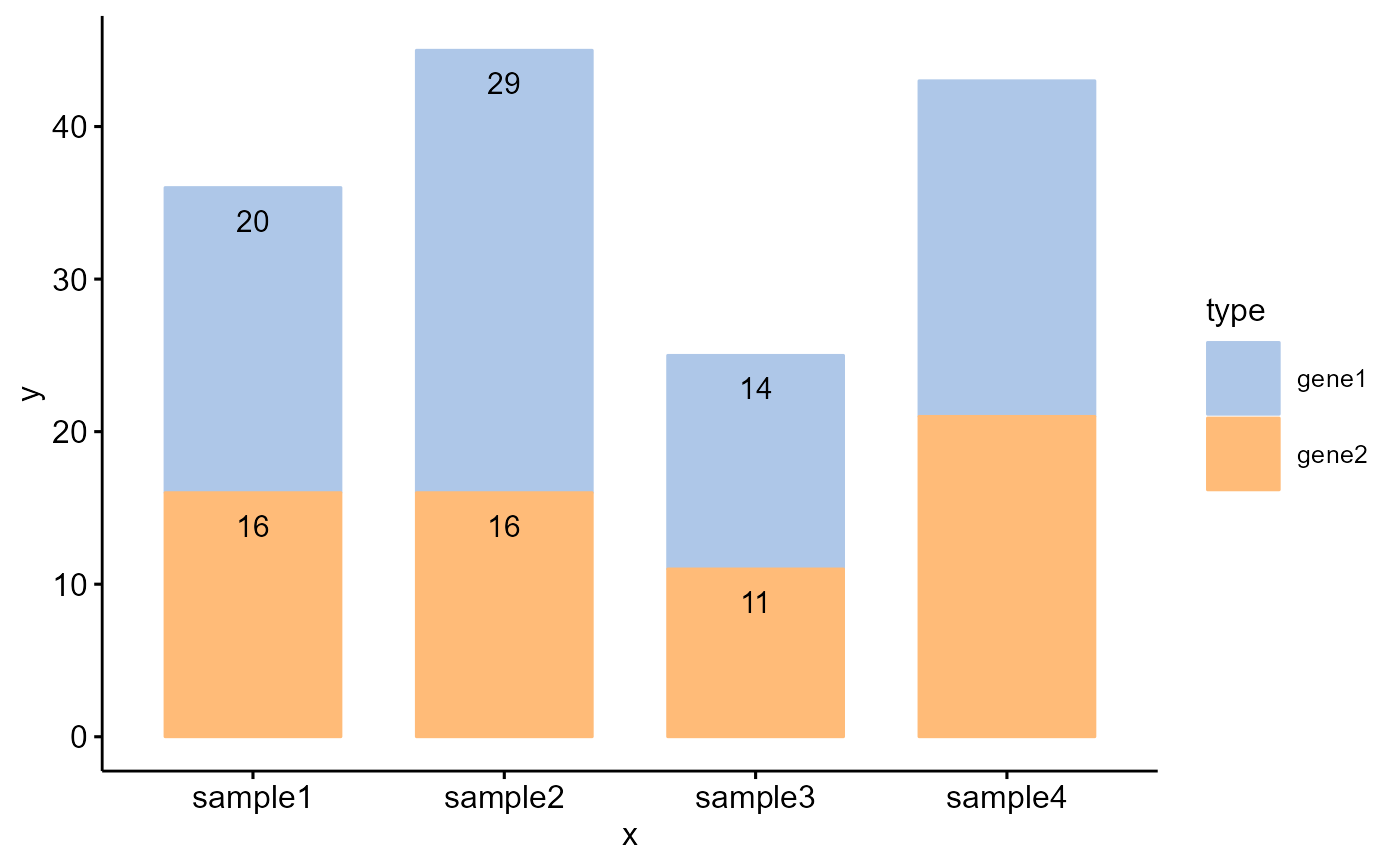 # Examples 3
Bior_BarPlot(df, "x", "y", fill = "type", color = "type", palette = col,
label = TRUE, position = position_dodge(0.9))
# Examples 3
Bior_BarPlot(df, "x", "y", fill = "type", color = "type", palette = col,
label = TRUE, position = position_dodge(0.9))