---
author: "Hu Zheng"
date: "2025-12-01"
date-format: YYYY-MM-DD
---
# FigureS4
```{r}
#| warning: false
#| message: false
library(Seurat)
library(tidyverse)
library(hdWGCNA)
library(scRNAtoolVis)
source('bin/Palettes.R')
```
```{r}
seu.harmony <- readRDS('../data/seu.harmony.rds')
seu.IT <- subset(seu.harmony, cells=colnames(seu.harmony)[which(
seu.harmony$SubType %in% names(col_SubType)[1:8]
)])
seu.Ex <- seu.harmony[,seu.harmony$SubType %in% names(col_SubType)[1:11]]
```
```{r}
seurat_obj <- readRDS("../data/Figure4/seu.Ex.hdwgcna.rds")
```
## Figure_S4A
```{r}
seu <- seu.IT
seu$Time_SubType <- paste(seu$orig.ident, seu$SubType)
Time_SubType <- c(
"P1 Im L2/3 IT","P1 Im L4/5 IT","P1 Im L5 IT","P1 Im L6 IT","P1 L6 IT",
"P4 Im L2/3 IT","P4 Im L4/5 IT","P4 Im L5 IT","P4 L5 IT","P4 Im L6 IT","P4 L6 IT",
"P10 Im L2/3 IT","P10 L2/3 IT","P10 Im L4/5 IT","P10 L4/5 IT","P10 Im L5 IT",
"P10 L5 IT","P10 Im L6 IT","P10 L6 IT",
"Adult L2/3 IT","Adult L4/5 IT","Adult L5 IT","Adult L6 IT"
)
seu <- seu[,seu$Time_SubType %in% Time_SubType]
seu$Time_SubType <- factor(seu$Time_SubType, levels = Time_SubType)
Idents(seu) <- "Time_SubType"
#DEGs <- FindAllMarkers(seu, features = all_gene)
#write.csv(DEGs, "../data/Figure4/DEGs_IT_Time_SubType.csv")
DEGs <- read.csv("../data/Figure4/DEGs_IT_Time_SubType.csv", row.names = 1)
```
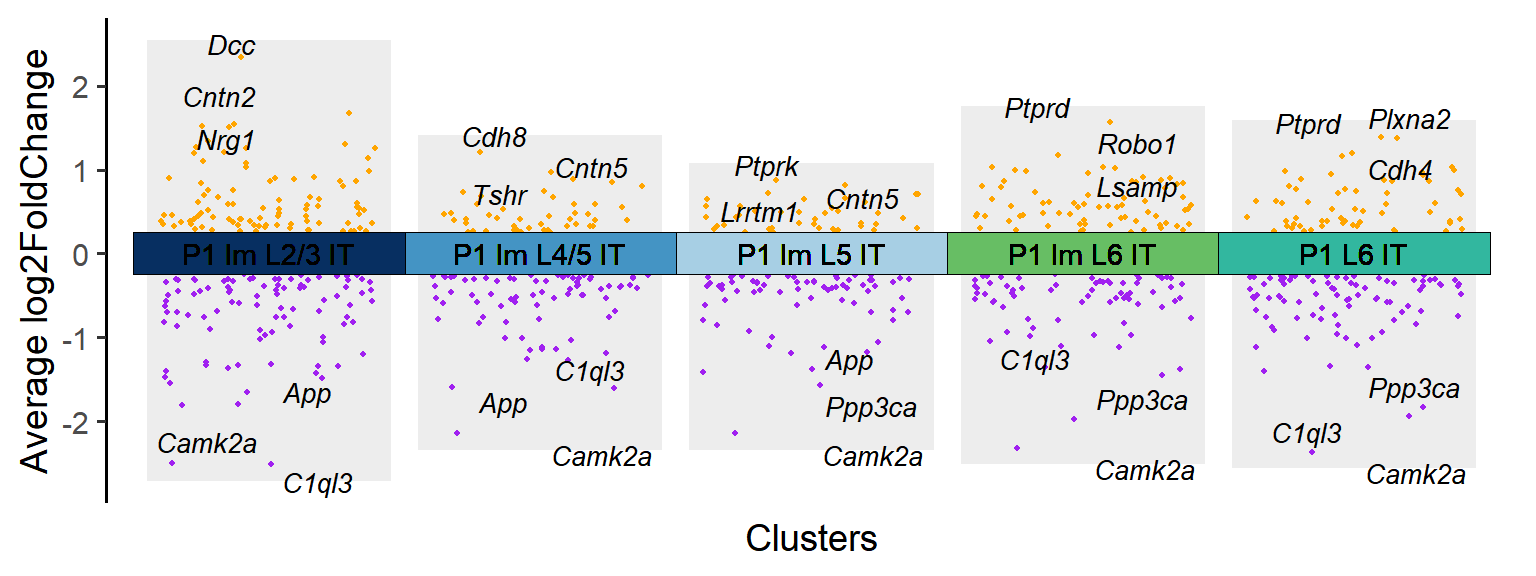
### Figure_S4A_1
```{r fig.width=8, fig.height=3}
#| message: false
P1_cluster <- Time_SubType[1:5]
P1_DEGs <- DEGs[DEGs$cluster %in% P1_cluster,]
tile.col <- c("P1 Im L2/3 IT"="#062e60","P1 Im L4/5 IT"="#4393c3","P1 Im L5 IT"="#a6cee3","P1 Im L6 IT"="#66bd63","P1 L6 IT"="#31b69e")
Figure_S4A_1 <- jjVolcano(
diffData = P1_DEGs,
aesCol = c('purple','orange'),
topGeneN = 3,
tile.col = tile.col,
size = 3.5, segment.size = 0.1,
fontface = 'italic'
) + NoLegend()
Figure_S4A_1
```
```{r}
#| eval: false
ggsave("../../Figure/FigureS4/Figure_S4A_1.pdf", plot = Figure_S4A_1,
height = 3, width = 8, units = "in")
```
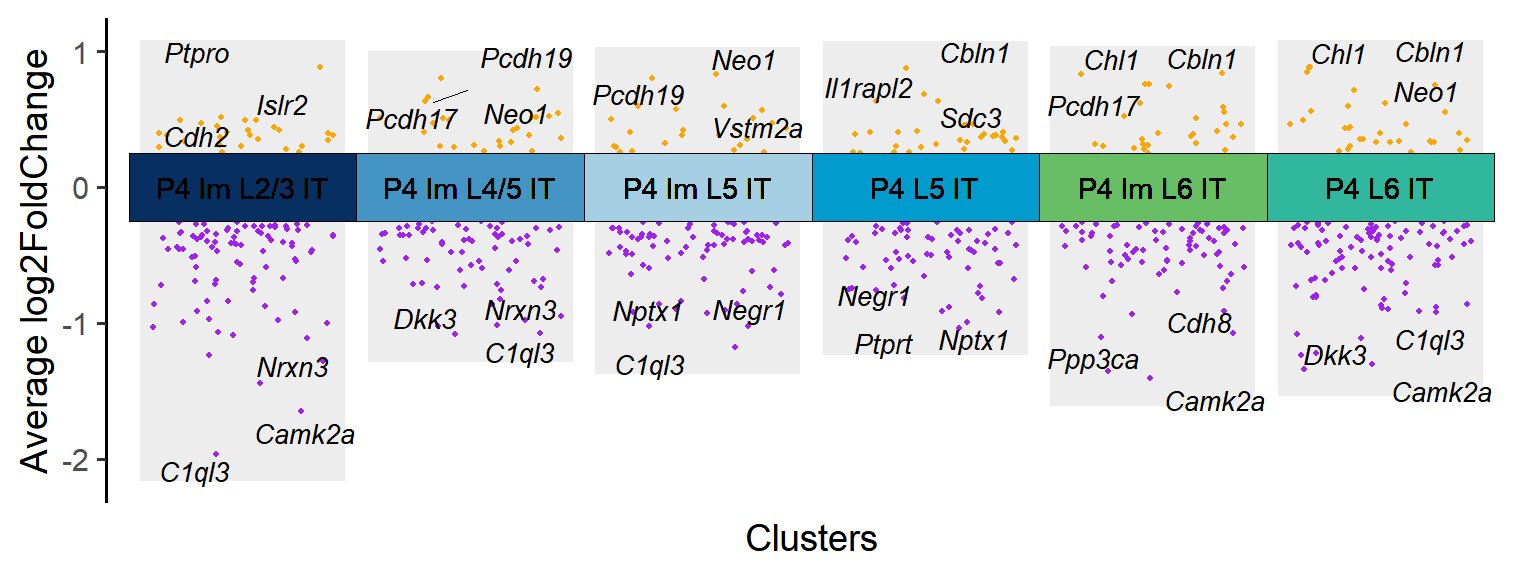
### Figure_S4A_2
```{r fig.width=8, fig.height=3}
#| message: false
P4_cluster <- Time_SubType[6:11]
P4_DEGs <- DEGs[DEGs$cluster %in% P4_cluster,]
tile.col <- c("P4 Im L2/3 IT"="#062e60","P4 Im L4/5 IT"="#4393c3","P4 Im L5 IT"="#a6cee3","P4 L5 IT"="#009acd","P4 Im L6 IT"="#66bd63","P4 L6 IT"="#31b69e")
Figure_S4A_2 <- jjVolcano(
diffData = P4_DEGs,
aesCol = c('purple','orange'),
topGeneN = 3,
tile.col = tile.col, cluster.order = names(tile.col),
size = 3.5, segment.size = 0.1,
fontface = 'italic'
) + NoLegend()
Figure_S4A_2
```
```{r}
#| eval: false
ggsave("../../Figure/FigureS4/Figure_S4A_2.pdf", plot = Figure_S4A_2,
height = 3, width = 8, units = "in")
```
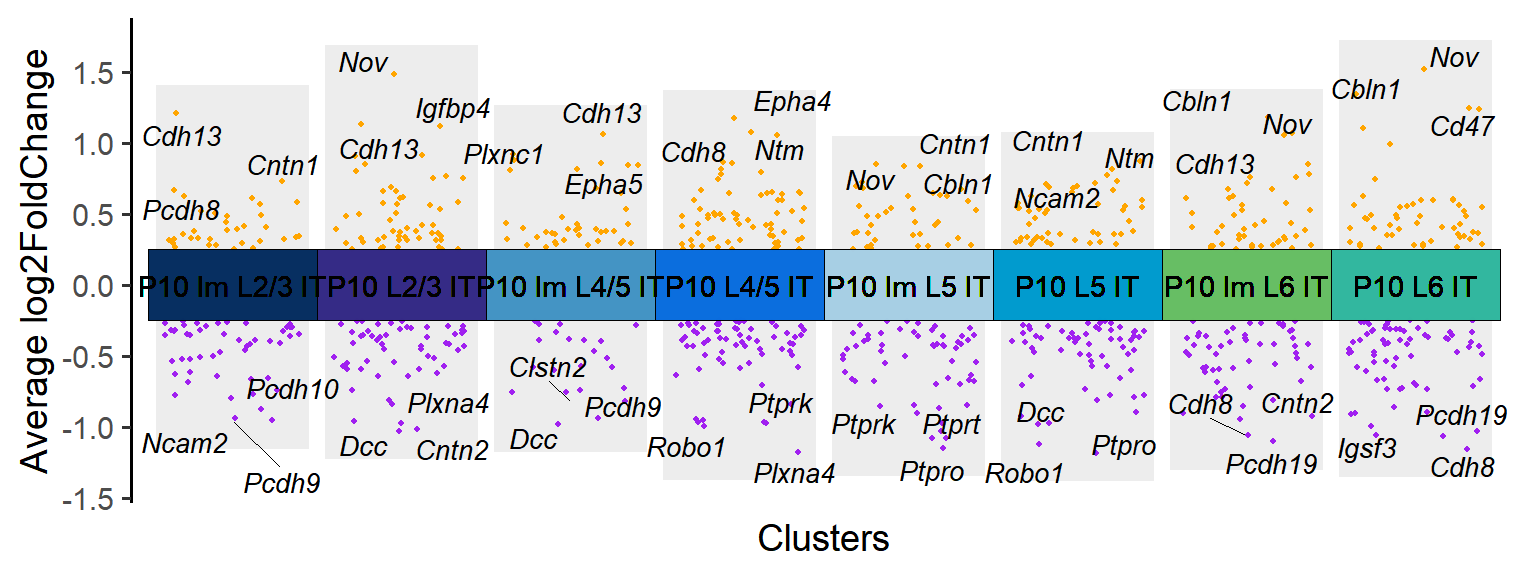
### Figure_S4A_3
```{r fig.width=8, fig.height=3}
#| message: false
P10_cluster <- Time_SubType[12:19]
P10_DEGs <- DEGs[DEGs$cluster %in% P10_cluster,]
tile.col <- c("P10 Im L2/3 IT"="#062e60","P10 L2/3 IT"="#342a85","P10 Im L4/5 IT"="#4393c3","P10 L4/5 IT"="#0a6ddd","P10 Im L5 IT"="#a6cee3","P10 L5 IT"="#009acd","P10 Im L6 IT"="#66bd63","P10 L6 IT"="#31b69e")
Figure_S4A_3 <- jjVolcano(
diffData = P10_DEGs,
aesCol = c('purple','orange'),
topGeneN = 3,
tile.col = tile.col, cluster.order = names(tile.col),
size = 3.5, segment.size = 0.1,
fontface = 'italic'
) + NoLegend()
Figure_S4A_3
```
```{r}
#| eval: false
ggsave("../../Figure/FigureS4/Figure_S4A_3.pdf", plot = Figure_S4A_3,
height = 3, width = 8, units = "in")
```
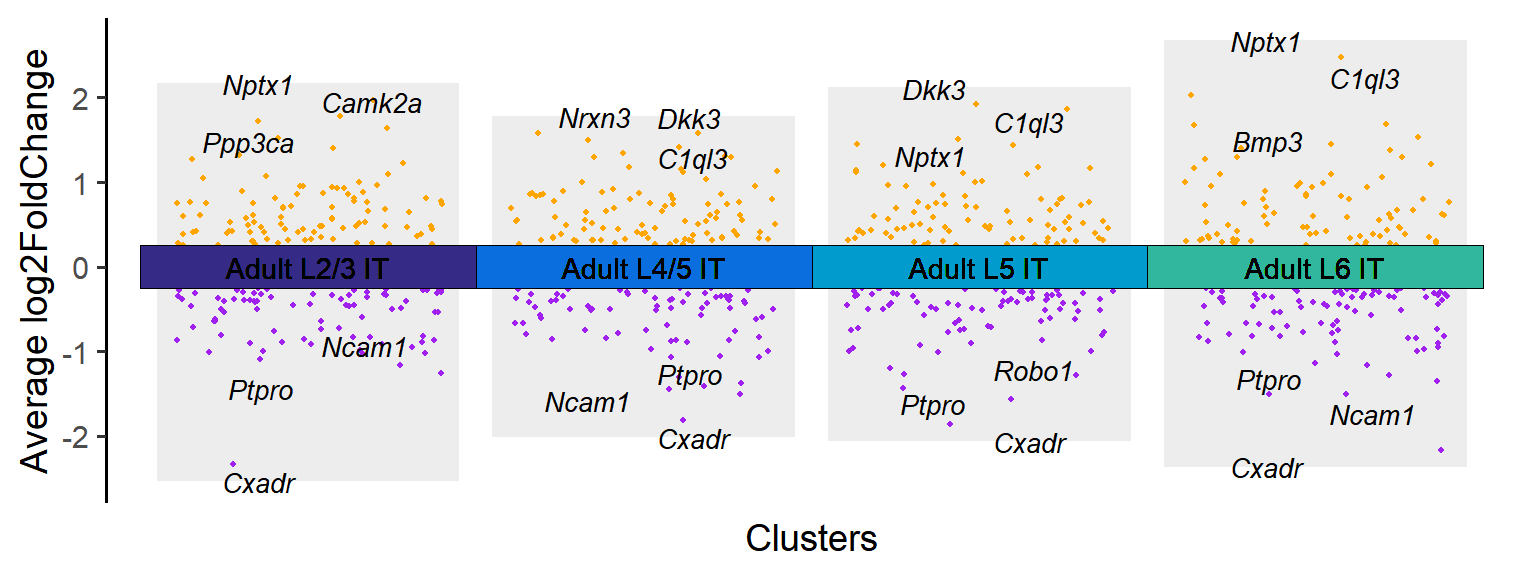
### Figure_S4A_4
```{r fig.width=8, fig.height=3}
#| message: false
Adult_cluster <- Time_SubType[20:23]
Adult_DEGs <- DEGs[DEGs$cluster %in% Adult_cluster,]
tile.col <- c("Adult L2/3 IT"="#342a85","Adult L4/5 IT"="#0a6ddd","Adult L5 IT"="#009acd","Adult L6 IT"="#31b69e")
Figure_S4A_4 <- jjVolcano(
diffData = Adult_DEGs,
aesCol = c('purple','orange'),
topGeneN = 3,
tile.col = tile.col, cluster.order = names(tile.col),
size = 3.5, segment.size = 0.1,
fontface = 'italic'
) + NoLegend()
Figure_S4A_4
```
```{r}
#| eval: false
ggsave("../../Figure/FigureS4/Figure_S4A_4.pdf", plot = Figure_S4A_4,
height = 3, width = 8, units = "in")
```
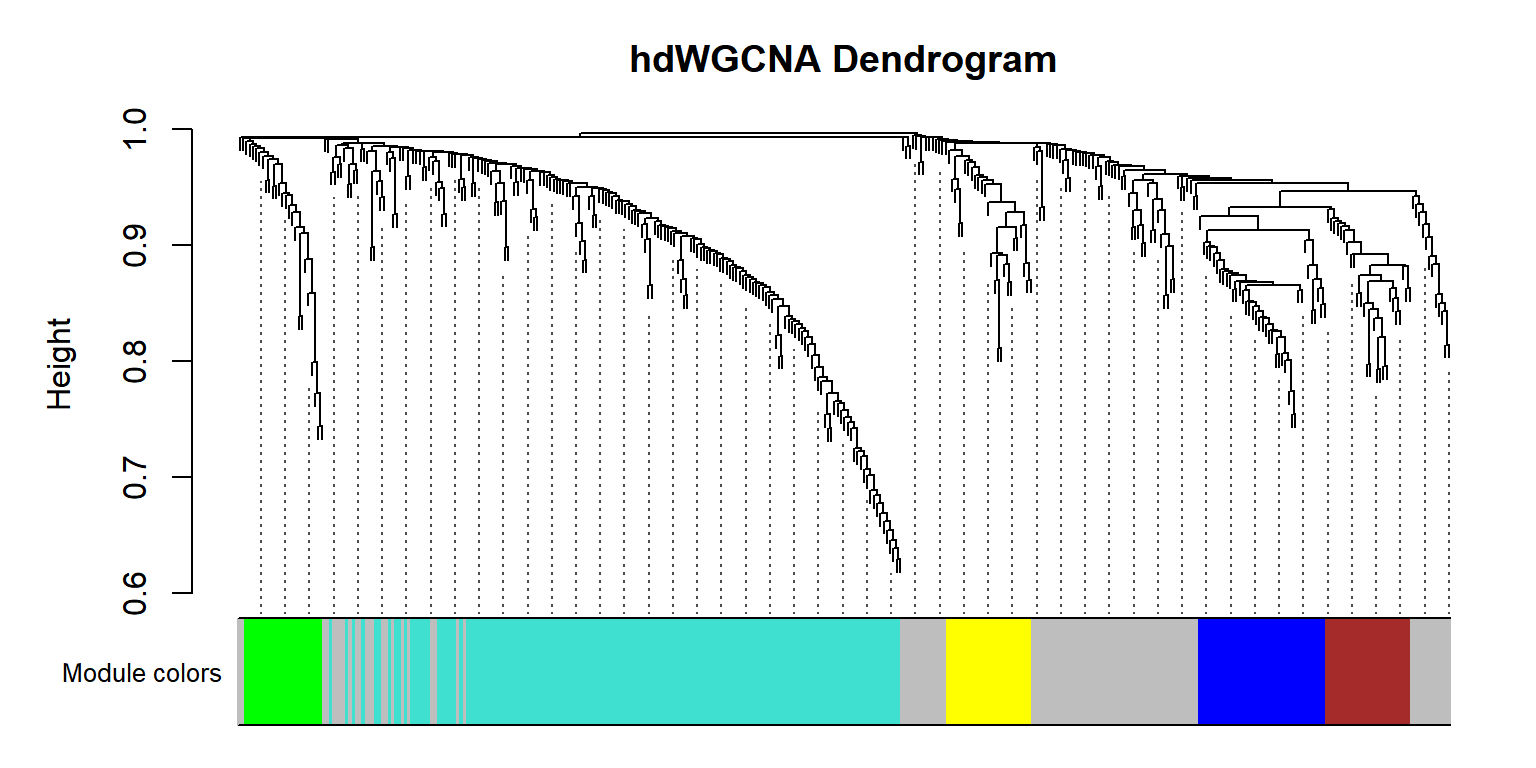
## Figure_S4B
```{r fig.width=8, fig.height=4}
#| message: false
#| warning: false
Figure_S4B <- PlotDendrogram(seurat_obj, main='hdWGCNA Dendrogram')
Figure_S4B
```
```{r}
#| eval: false
ggsave("../../Figure/FigureS4/Figure_S4B.pdf", plot = Figure_S4B,
height = 4, width = 8, units = "in")
```
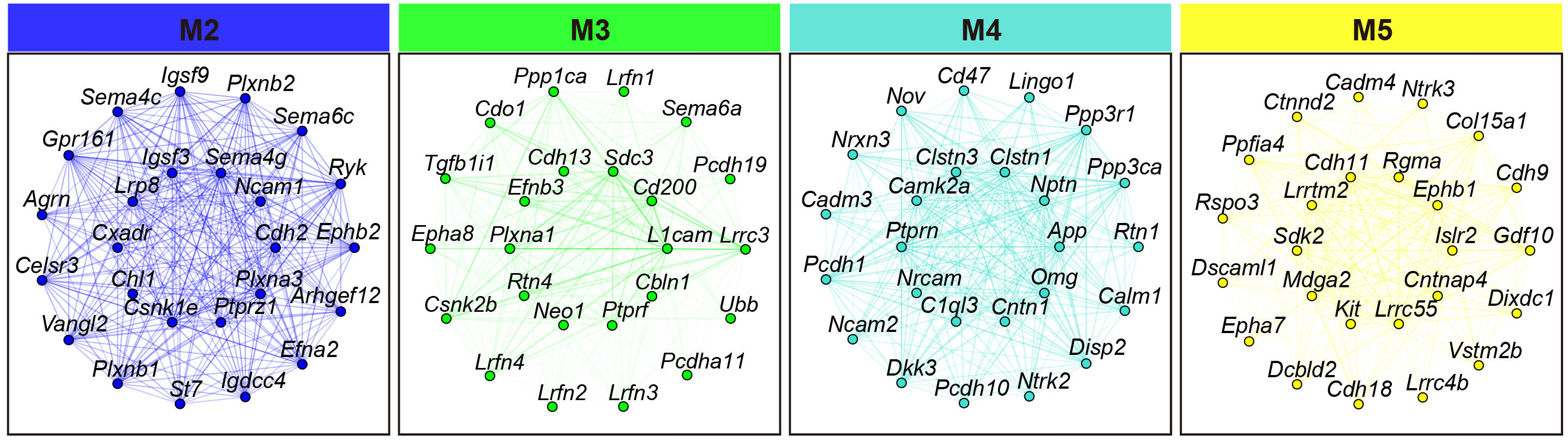
## Figure_S4C
```{r}
#| eval: false
ModuleNetworkPlot(
seurat_obj,
outdir = '../../Figure/Figure4/ModuleNetwork'
)
```
```{r}
knitr::include_graphics("./images/Figure_S4C.jpg", dpi = 300)
```
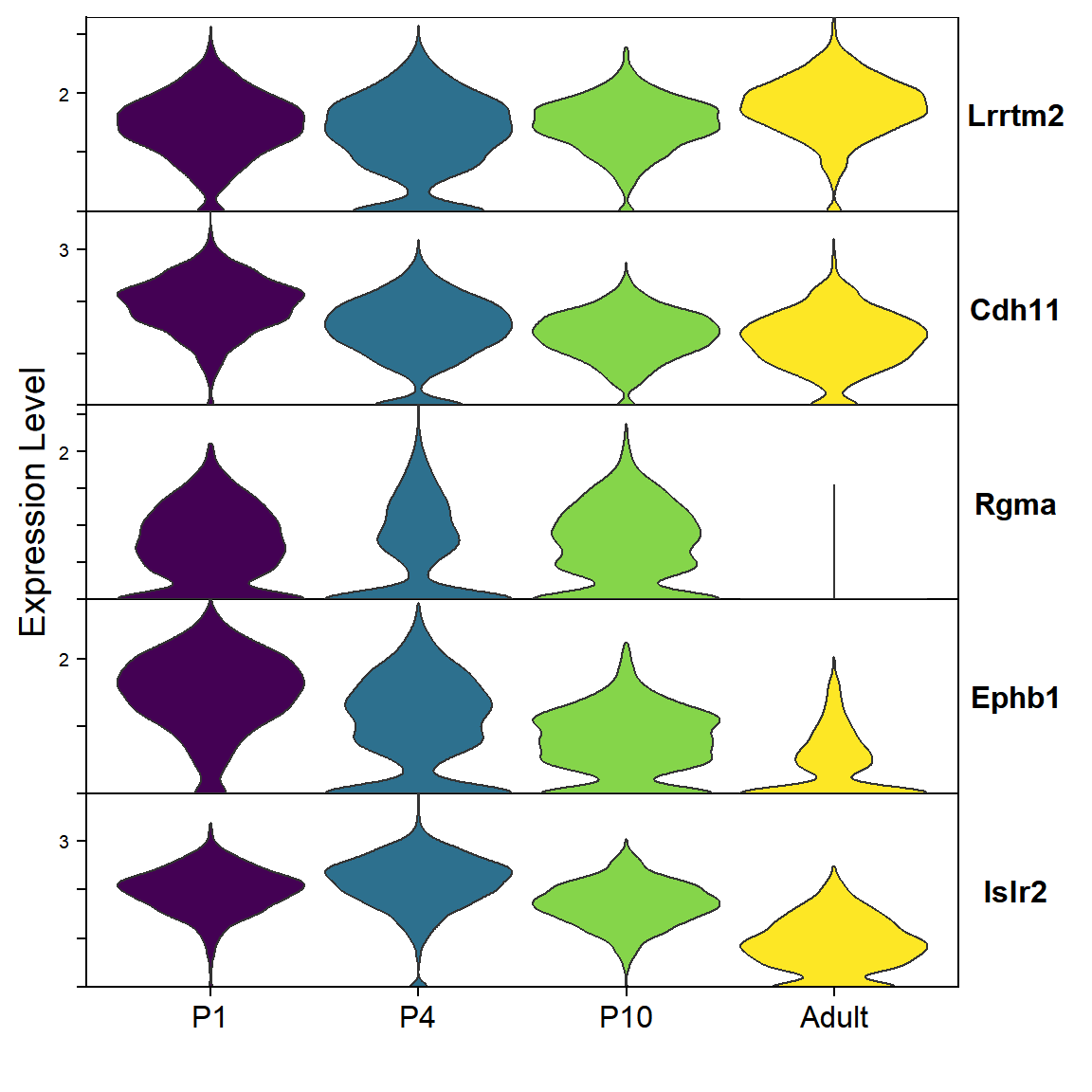
## Figure_S4D
```{r fig.width=6, fig.height=6}
#| message: false
#seu <- seu.IT
seu <- seu.Ex[,seu.Ex$SubType=="L6 CT"]
seu$orig.ident <- factor(seu$orig.ident, levels = c("P1","P4","P10","Adult"))
#M2
#gene_list <- c("Cdh2","Ncam1","Sema4g","Igsf3","Lrp8")
#M3
#gene_list <- c("Efnb3","Cdh13","Sdc3","Cd200","L1cam")
#M4
#gene_list <- c("App","Nptn","Clstn1","Clstn3","Camk2a")
#M5
gene_list <- c("Islr2","Ephb1","Rgma","Cdh11","Lrrtm2")
Figure_S4D <-
VlnPlot(seu, features = rev(gene_list), group.by = "orig.ident", cols=col_Time,
split.by = "orig.ident",
stack = TRUE, flip = TRUE) +
theme(legend.position = "none", axis.text.x = element_text(angle = 0, hjust = 0.5)) +
labs(x="")
Figure_S4D
```
```{r}
#| eval: false
ggsave("../../Figure/FigureS4/Figure_S4D_M5.pdf", plot = Figure_S4D,
height = 6, width = 6, units = "in")
```
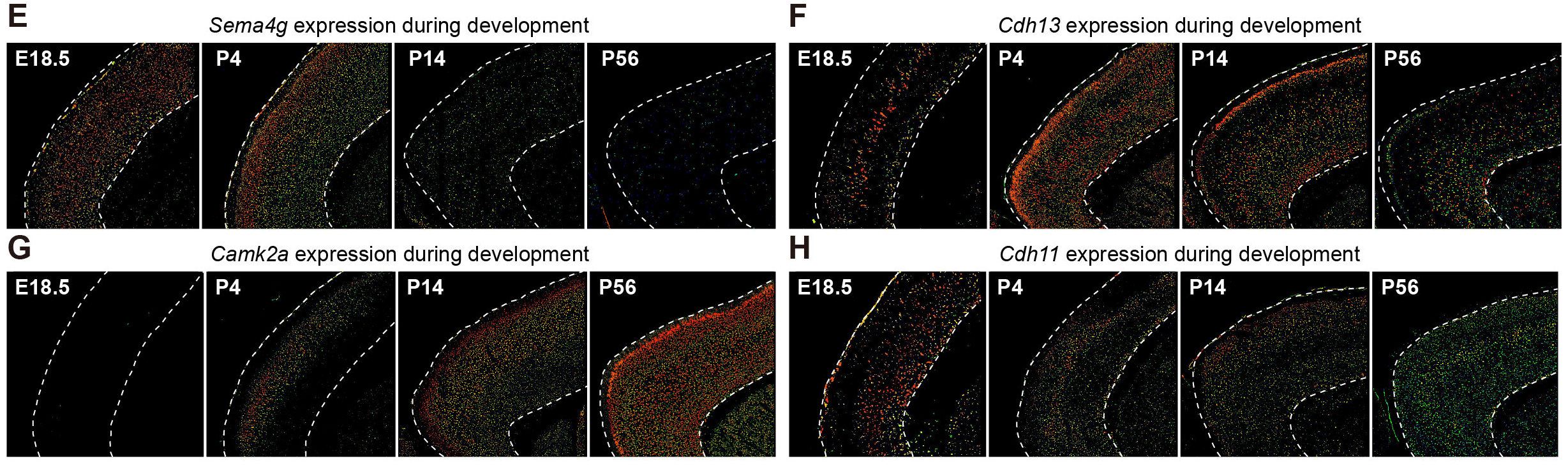
## Figure_S4E-H
```{r}
knitr::include_graphics("./images/Figure_S4E-H.png", dpi = 300)
```