---
author: "Hu Zheng"
date: "2025-12-01"
date-format: YYYY-MM-DD
---
# FigureS6
```{r}
#| warning: false
#| message: false
library(Seurat)
library(tidyverse)
library(scCustomize)
source('bin/Palettes.R')
```
```{r}
seu.harmony <- readRDS('../data/seu.harmony.rds')
```
```{r}
seu <- seu.harmony
seu$Time_Subtype <- paste(seu$orig.ident, seu$SubType)
seu <- seu[,!seu$Time_Subtype %in% c(
"P1 L2/3 IT","P1 L4/5 IT","P1 L5 IT","P1 Endo",
"P4 L2/3 IT","P4 L4/5 IT","P4 Endo","P10 Endo",
"Adult Im L2/3 IT","Adult Im L4/5 IT","Adult Im L5 IT","Adult Im L6 IT",
"Adult Endo","Adult NPC")]
```
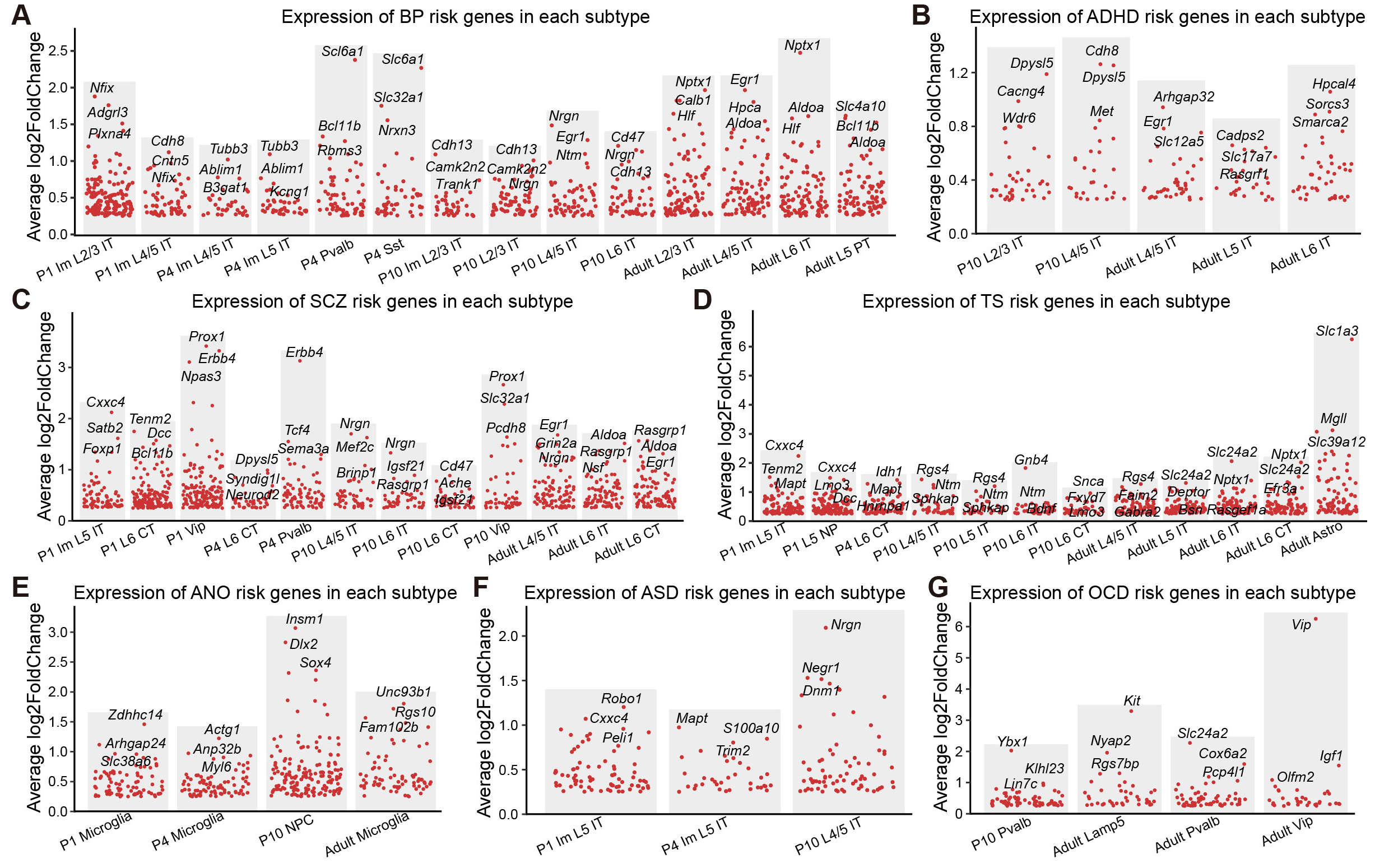
## Figure_S6A-G
```{r}
ADHD_subtype <- c("P10 L2/3 IT","P10 L4/5 IT","Adult L4/5 IT","Adult L5 IT",
"Adult L6 IT")
ANO_subtype <- c("P1 Microglia","P4 Microglia","P10 NPC","Adult Microglia")
ASD_subtype <- c("P1 Im L5 IT","P4 Im L5 IT","P10 L4/5 IT")
BP_subtype <- c("P1 Im L2/3 IT","P1 Im L4/5 IT","P4 Im L4/5 IT","P4 Im L5 IT",
"P4 Pvalb","P4 Sst","P10 Im L2/3 IT","P10 L2/3 IT","P10 L4/5 IT",
"P10 L6 IT","Adult L2/3 IT","Adult L4/5 IT","Adult L6 IT",
"Adult L5 PT")
MDD_subtype <- c("P1 Im L6 IT","P1 Lamp5","P1 Pvalb","P1 Vip","P4 L6 IT",
"P4 Pvalb","P4 Vip","P10 Im L6 IT","P10 L2/3 IT","P10 L4/5 IT",
"P10 L5 IT","P10 L6 IT","P10 Lamp5","P10 Pvalb","P10 Vip",
"Adult L2/3 IT","Adult L4/5 IT","Adult L5 IT","Adult L6 IT",
"Adult L5 NP","Adult L6 CT","Adult Pvalb","Adult Vip")
OCD_subtype <- c("P10 Pvalb","Adult Lamp5","Adult Pvalb","Adult Vip")
SCZ_subtype <- c("P1 Im L5 IT","P1 L6 CT","P1 Vip","P4 L6 CT","P4 Pvalb",
"P10 L4/5 IT","P10 L6 IT","P10 L6 CT","P10 Vip","Adult L4/5 IT",
"Adult L6 IT","Adult L6 CT")
TS_subtype <- c("P1 Im L5 IT","P1 L5 NP","P4 L6 CT","P10 L4/5 IT","P10 L5 IT",
"P10 L6 IT","P10 L6 CT","Adult L4/5 IT","Adult L5 IT",
"Adult L6 IT","Adult L6 CT","Adult Astro")
Gene_weight <- read.csv("../../data/Revision/Gene_weight.csv")
```
```{r fig.width=10, fig.height=3}
#| eval: false
disease_subtype <- BP_subtype
gene <- Gene_weight$BP_Gene
seu_disease <- seu[,seu$Time_Subtype %in% disease_subtype]
gene <- gene[gene %in% rownames(seu)]
seu_disease$Time_Subtype <- factor(seu_disease$Time_Subtype,
levels = disease_subtype)
Idents(seu_disease) <- "Time_Subtype"
DEG <- FindAllMarkers(seu_disease, features = gene, logfc.threshold = 0.25,
only.pos = T)
Figure_S6A <- jjVolcano(
diffData = DEG,
topGeneN = 0,
log2FC.cutoff = 0.1,
col.type = "updown",
tile.col = rep("lightgray",34),
aesCol = c('#0099CC','#CC3333'),
size = 5,
fontface = 'italic'
)
Figure_S6A
```
```{r}
knitr::include_graphics("./images/Figure_S6A-G.png", dpi = 300)
```
```{r}
#| eval: false
ggsave("../../Figure/FigureS6/Figure_S6A_BP.pdf", plot = Figure_6B,
height = 3, width = 10, units = "in")
```
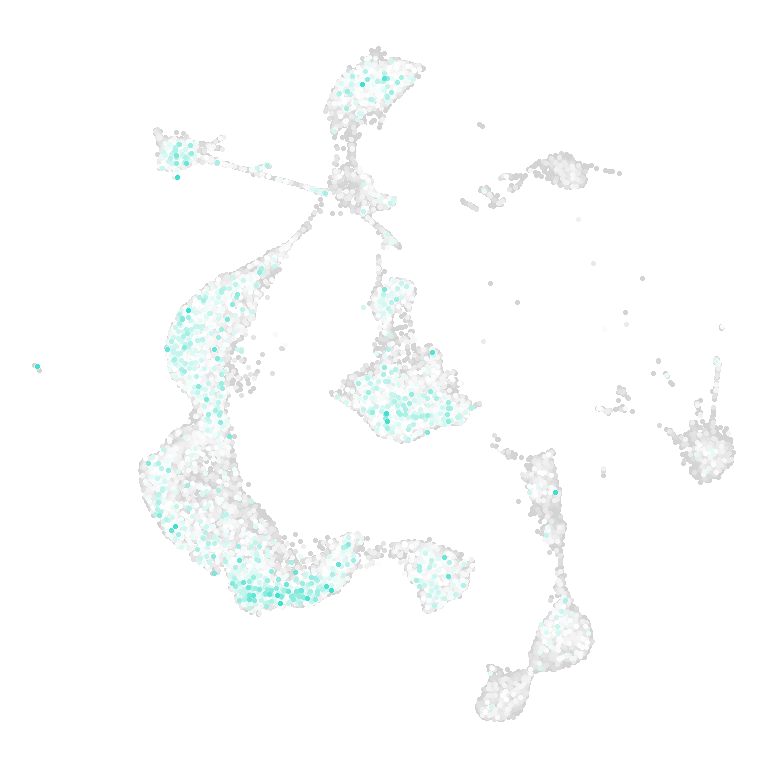
## Figure_S6H
```{r}
P1_score <- read.csv("../../data/Revision/P1/P1_score.csv", row.names = 1)
P4_score <- read.csv("../../data/Revision/P4/P4_score.csv", row.names = 1)
P10_score <- read.csv("../../data/Revision/P10/P10_score.csv", row.names = 1)
Adult_score <- read.csv("../../data/Revision/Adult/Adult_score.csv", row.names = 1)
All_score <- rbind(P1_score, P4_score, P10_score, Adult_score)
```
```{r fig.width=4, fig.height=4}
disease <- names(col_Disease)[1]
df <- data.frame(
UMAP_1 = All_score$UMAP_1,
UMAP_2 = All_score$UMAP_2,
value = All_score[,disease]
)
df$value[df$value > 4] <- 4
df$value[df$value < 0] <- 0
df <- df[order(df$value),]
Figure_S6H <-
ggplot() +
geom_point(df, mapping=aes(x=UMAP_1, y=UMAP_2, color=value), size=0.5) +
theme_void() +
theme(legend.position = "none") +
coord_fixed() +
scale_color_gradientn(colours = c("lightgray", "white", col_Disease[1]),
limits = c(0,4))
Figure_S6H
```
```{r}
#| eval: false
ggsave(filename=paste("../../Figure/FigureS6/Figure_S6H/",disease,".png",sep=""),
Figure_6C, width=4, height = 4, units = "in")
```