---
author: "Hu Zheng"
date: "2025-12-01"
date-format: YYYY-MM-DD
---
# Figure6
```{r}
#| warning: false
#| message: false
library(Seurat)
library(tidyverse)
library(stringr)
library(ggplot2)
library(pheatmap)
library(cowplot)
library(ggalt)
library(scRNAtoolVis)
library(scCustomize)
source('bin/Palettes.R')
```
```{r}
seu.harmony <- readRDS('../data/seu.harmony.rds')
```
```{r}
seu <- seu.harmony
seu$Time_Subtype <- paste(seu$orig.ident, seu$SubType)
seu <- seu[,!seu$Time_Subtype %in% c(
"P1 L2/3 IT","P1 L4/5 IT","P1 L5 IT","P1 Endo",
"P4 L2/3 IT","P4 L4/5 IT","P4 Endo","P10 Endo",
"Adult Im L2/3 IT","Adult Im L4/5 IT","Adult Im L5 IT","Adult Im L6 IT",
"Adult Endo","Adult NPC")]
```
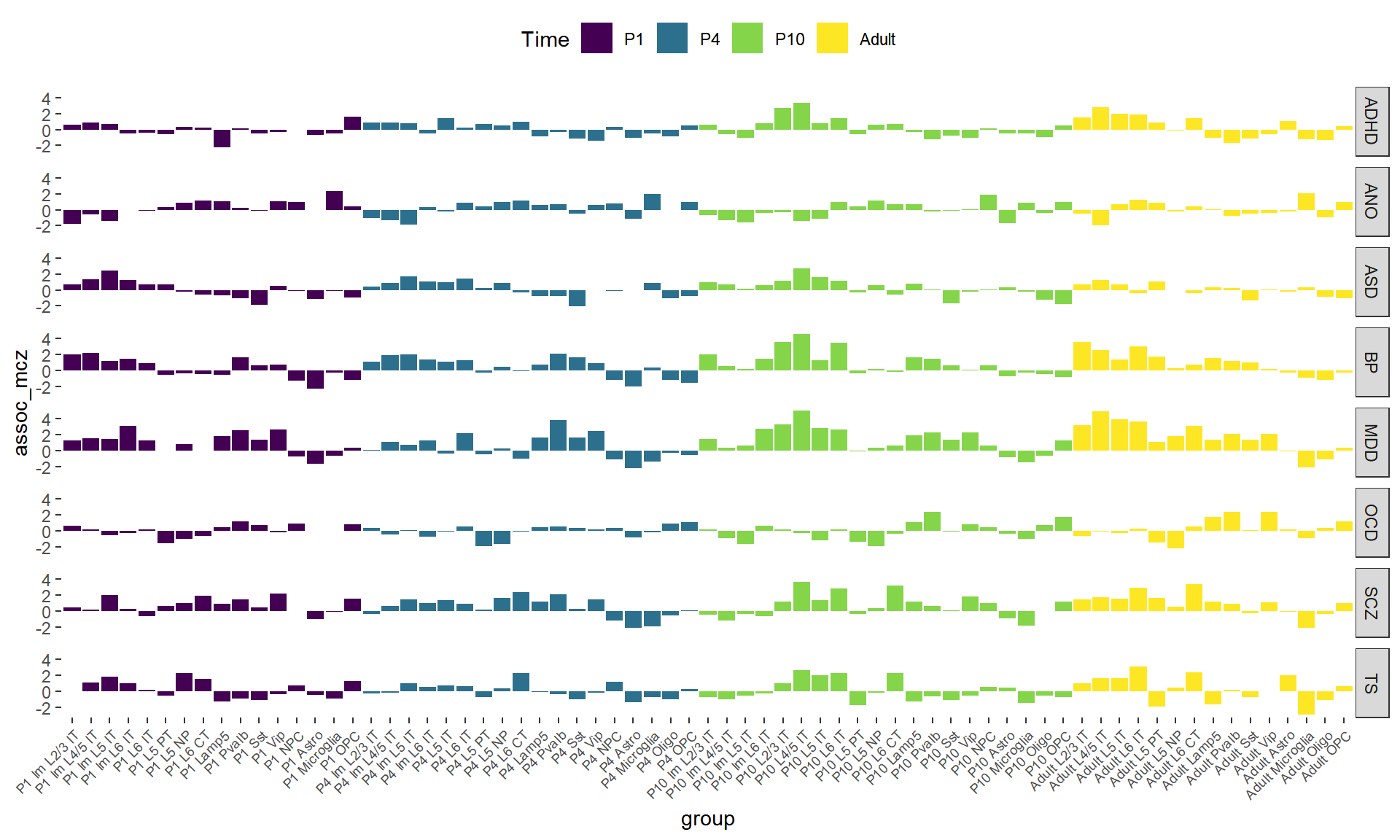
## Figure_6A
```{r fig.width=10, fig.height=6}
All_Disease_separate <- read.csv("../../data/Revision/All_Disease_separate.csv")
disease_result <- All_Disease_separate
order <- paste(rep(c("P1","P4","P10","Adult"), each=21),
rep(names(col_SubType),4))
order <- order[order %in% disease_result$group]
disease_result$group <- factor(disease_result$group, levels = order)
disease_result$Time <- factor(disease_result$Time, levels = c("P1","P4","P10","Adult"))
Figure_6A <-
ggplot(data = disease_result, mapping = aes(x = group, y = assoc_mcz, fill = Time)) +
geom_bar(stat = 'identity') +
facet_grid(Disease ~ .) +
scale_fill_manual(values = col_Time) +
theme_bw() +
theme(axis.text.x = element_text(angle = 45, hjust = 1, size = 7),
panel.grid = element_blank(), panel.border = element_blank(),
legend.position = "top")
Figure_6A
```
```{r}
#| eval: false
ggsave("../../Figure/Figure6/Figure_6A.pdf", plot = Figure_6A,
height = 6, width = 10, units = "in")
```
## Figure_6B
```{r}
ADHD_subtype <- c("P10 L2/3 IT","P10 L4/5 IT","Adult L4/5 IT","Adult L5 IT",
"Adult L6 IT")
ANO_subtype <- c("P1 Microglia","P4 Microglia","P10 NPC","Adult Microglia")
ASD_subtype <- c("P1 Im L5 IT","P4 Im L5 IT","P10 L4/5 IT")
BP_subtype <- c("P1 Im L2/3 IT","P1 Im L4/5 IT","P4 Im L4/5 IT","P4 Im L5 IT",
"P4 Pvalb","P4 Sst","P10 Im L2/3 IT","P10 L2/3 IT","P10 L4/5 IT",
"P10 L6 IT","Adult L2/3 IT","Adult L4/5 IT","Adult L6 IT",
"Adult L5 PT")
MDD_subtype <- c("P1 Im L6 IT","P1 Lamp5","P1 Pvalb","P1 Vip","P4 L6 IT",
"P4 Pvalb","P4 Vip","P10 Im L6 IT","P10 L2/3 IT","P10 L4/5 IT",
"P10 L5 IT","P10 L6 IT","P10 Lamp5","P10 Pvalb","P10 Vip",
"Adult L2/3 IT","Adult L4/5 IT","Adult L5 IT","Adult L6 IT",
"Adult L5 NP","Adult L6 CT","Adult Pvalb","Adult Vip")
OCD_subtype <- c("P10 Pvalb","Adult Lamp5","Adult Pvalb","Adult Vip")
SCZ_subtype <- c("P1 Im L5 IT","P1 L6 CT","P1 Vip","P4 L6 CT","P4 Pvalb",
"P10 L4/5 IT","P10 L6 IT","P10 L6 CT","P10 Vip","Adult L4/5 IT",
"Adult L6 IT","Adult L6 CT")
TS_subtype <- c("P1 Im L5 IT","P1 L5 NP","P4 L6 CT","P10 L4/5 IT","P10 L5 IT",
"P10 L6 IT","P10 L6 CT","Adult L4/5 IT","Adult L5 IT",
"Adult L6 IT","Adult L6 CT","Adult Astro")
Gene_weight <- read.csv("../../data/Revision/Gene_weight.csv")
```
```{r fig.width=10, fig.height=3}
#| eval: false
disease_subtype <- MDD_subtype
gene <- Gene_weight$MDD_Gene
seu_disease <- seu[,seu$Time_Subtype %in% disease_subtype]
gene <- gene[gene %in% rownames(seu)]
seu_disease$Time_Subtype <- factor(seu_disease$Time_Subtype,
levels = disease_subtype)
Idents(seu_disease) <- "Time_Subtype"
DEG <- FindAllMarkers(seu_disease, features = gene, logfc.threshold = 0.25,
only.pos = T)
Figure_6B <- jjVolcano(
diffData = DEG,
topGeneN = 0,
log2FC.cutoff = 0.1,
col.type = "updown",
tile.col = rep("lightgray",34),
aesCol = c('#0099CC','#CC3333'),
size = 5,
fontface = 'italic'
)
Figure_6B
```
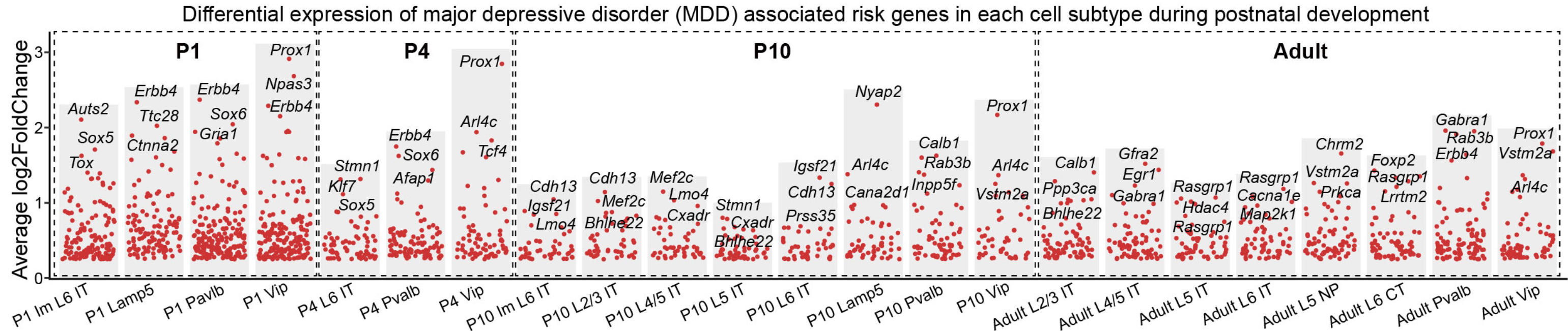
```{r}
knitr::include_graphics("./images/Figure_6B.png", dpi = 300)
```
```{r}
#| eval: false
ggsave("../../Figure/Figure6/Figure_6B_MDD.pdf", plot = Figure_6B,
height = 3, width = 10, units = "in")
```
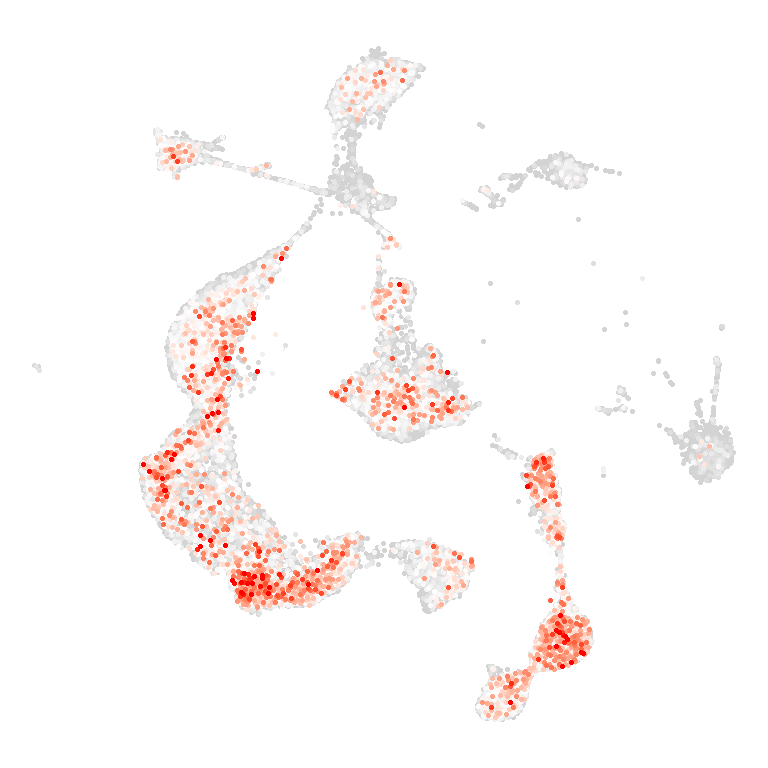
## Figure_6C
```{r}
P1_score <- read.csv("../../data/Revision/P1/P1_score.csv", row.names = 1)
P4_score <- read.csv("../../data/Revision/P4/P4_score.csv", row.names = 1)
P10_score <- read.csv("../../data/Revision/P10/P10_score.csv", row.names = 1)
Adult_score <- read.csv("../../data/Revision/Adult/Adult_score.csv", row.names = 1)
All_score <- rbind(P1_score, P4_score, P10_score, Adult_score)
```
```{r fig.width=4, fig.height=4}
disease <- names(col_Disease)[5]
df <- data.frame(
UMAP_1 = All_score$UMAP_1,
UMAP_2 = All_score$UMAP_2,
value = All_score[,disease]
)
df$value[df$value > 4] <- 4
df$value[df$value < 0] <- 0
df <- df[order(df$value),]
Figure_6C <-
ggplot() +
geom_point(df, mapping=aes(x=UMAP_1, y=UMAP_2, color=value), size=0.5) +
theme_void() +
theme(legend.position = "none") +
coord_fixed() +
scale_color_gradientn(colours = c("lightgray", "white", col_Disease[5]),
limits = c(0,4))
Figure_6C
```
```{r}
#| eval: false
ggsave(filename=paste("../../Figure/Figure6/Figure_6C/",disease,".png",sep=""),
Figure_6C, width=4, height = 4, units = "in")
```
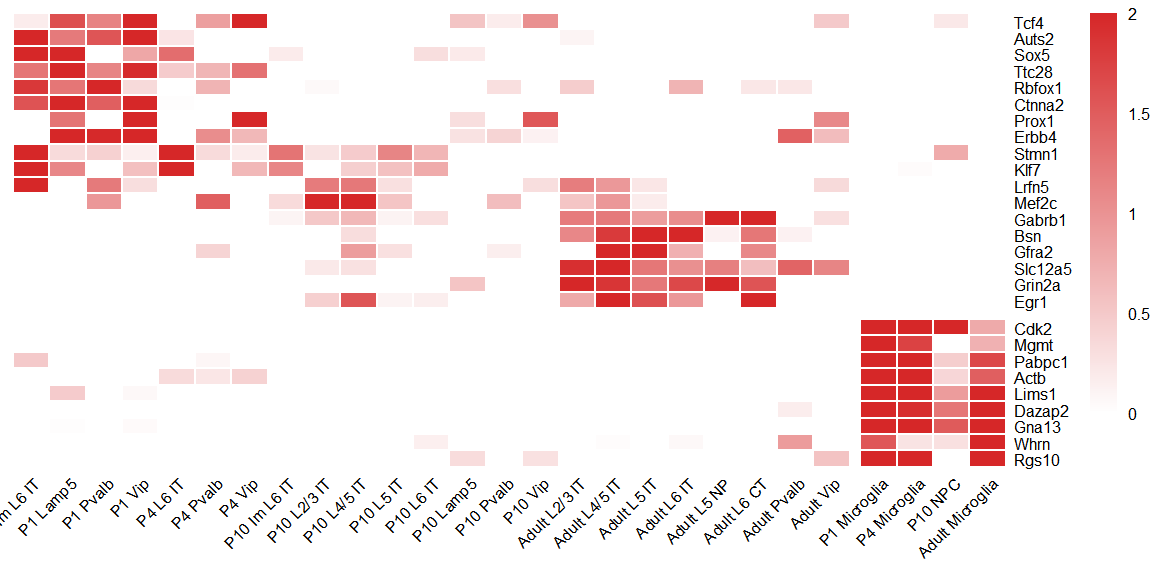
## Figure_6D
```{r fig.width=6, fig.height=3}
use_subtype <- c(order[order %in% MDD_subtype], order[order %in% ANO_subtype])
seu_heatmap <- seu[,seu$Time_Subtype %in% use_subtype]
seu_heatmap$Time_Subtype <- factor(seu_heatmap$Time_Subtype, levels = use_subtype)
MDD_gene <- c("Tcf4","Auts2","Sox5","Ttc28","Rbfox1","Ctnna2",
"Prox1","Erbb4","Stmn1","Klf7","Lrfn5","Mef2c",
"Gabrb1","Bsn","Gfra2","Slc12a5","Grin2a","Egr1")
ANO_gene <- c("Cdk2","Mgmt","Pabpc1","Actb","Lims1","Dazap2",
"Gna13","Whrn","Rgs10")
use_gene <- c(MDD_gene,ANO_gene)
avg_exp <- AverageExpression(seu_heatmap, group.by = "Time_Subtype", features = use_gene)$RNA
avg_exp_scale <- t(scale(t(avg_exp)))
avg_exp_scale[avg_exp_scale<0] <- 0
avg_exp_scale[avg_exp_scale>2] <- 2
Figure_6D <-
pheatmap(avg_exp_scale,
color = colorRampPalette(c("white","#d62728"))(200),
breaks = seq(0,2,0.01), show_colnames = T, treeheight_row=10,
cluster_rows = F, cluster_cols = F,
annotation_legend=F, fontsize = 6, angle_col=45,
gaps_col = 23, gaps_row = 18, border_color="white"
)
```
## Figure_6E
```{r fig.width=5, fig.height=5}
#| message: false
MDD_subtype <- c("P1 Im L6 IT","P1 Lamp5","P1 Pvalb","P1 Vip","P4 L6 IT",
"P4 Pvalb","P4 Vip","P10 Im L6 IT","P10 L2/3 IT","P10 L4/5 IT",
"P10 L5 IT","P10 L6 IT","P10 Lamp5","P10 Pvalb","P10 Vip",
"Adult L2/3 IT","Adult L4/5 IT","Adult L5 IT","Adult L6 IT",
"Adult L5 NP","Adult L6 CT","Adult Pvalb","Adult Vip")
seu <- seu.harmony[,seu.harmony$SubType %in% names(col_SubType)[1:8]]
seu$Time_Subtype <- paste(seu$orig.ident, seu$SubType)
seu <- seu[,seu$Time_Subtype %in% MDD_subtype]
seu$orig.ident <- factor(seu$orig.ident, levels = c("P1","P4","P10","Adult"))
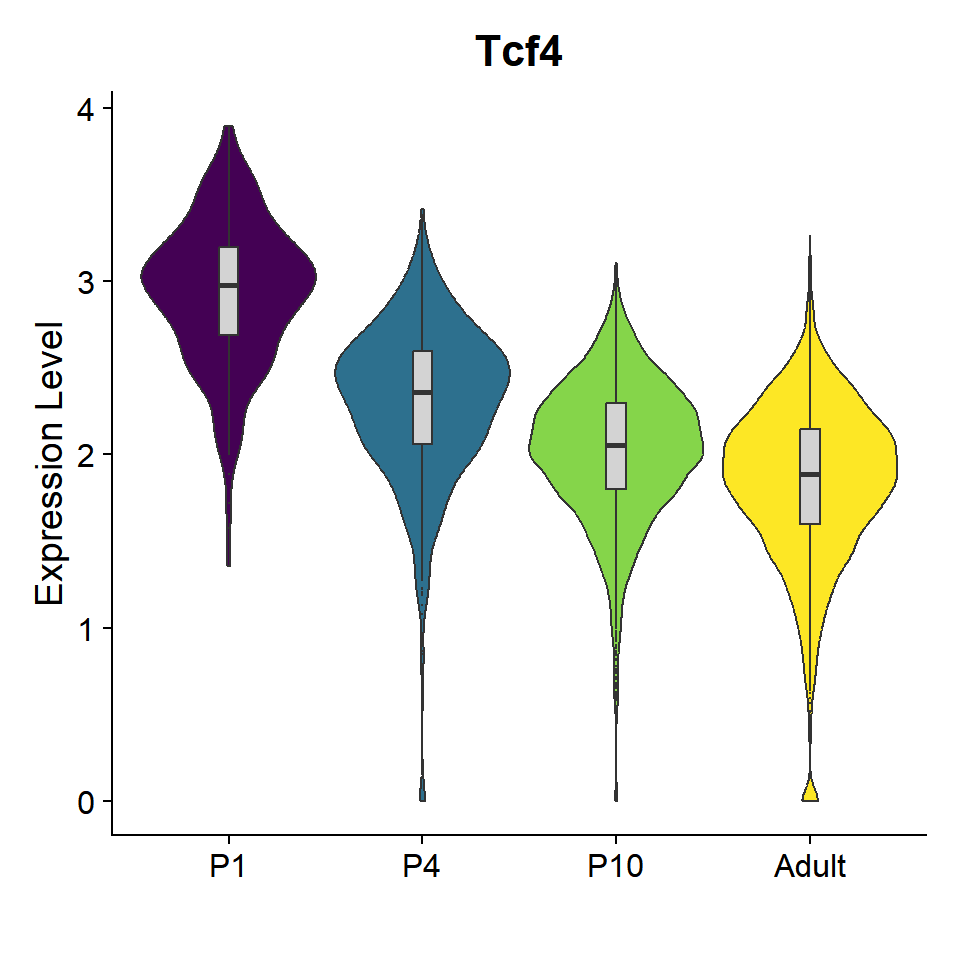
gene_list <- c("Tcf4")
Figure_6E <-
VlnPlot_scCustom(seurat_object = seu, features = gene_list, group.by = "orig.ident",
pt.size = 0) & NoLegend() &
geom_boxplot(width=0.1, outlier.size=0, fill="lightgray") &
scale_fill_manual(values = col_Time) &
theme(axis.text.x = element_text(angle = 0, hjust = 0.5)) &
labs(x="")
Figure_6E
```
```{r}
#| eval: false
ggsave("../../Figure/Figure6/Figure_6E_Tcf4.pdf", plot = Figure_6E,
height = 6, width = 5, units = "in")
```
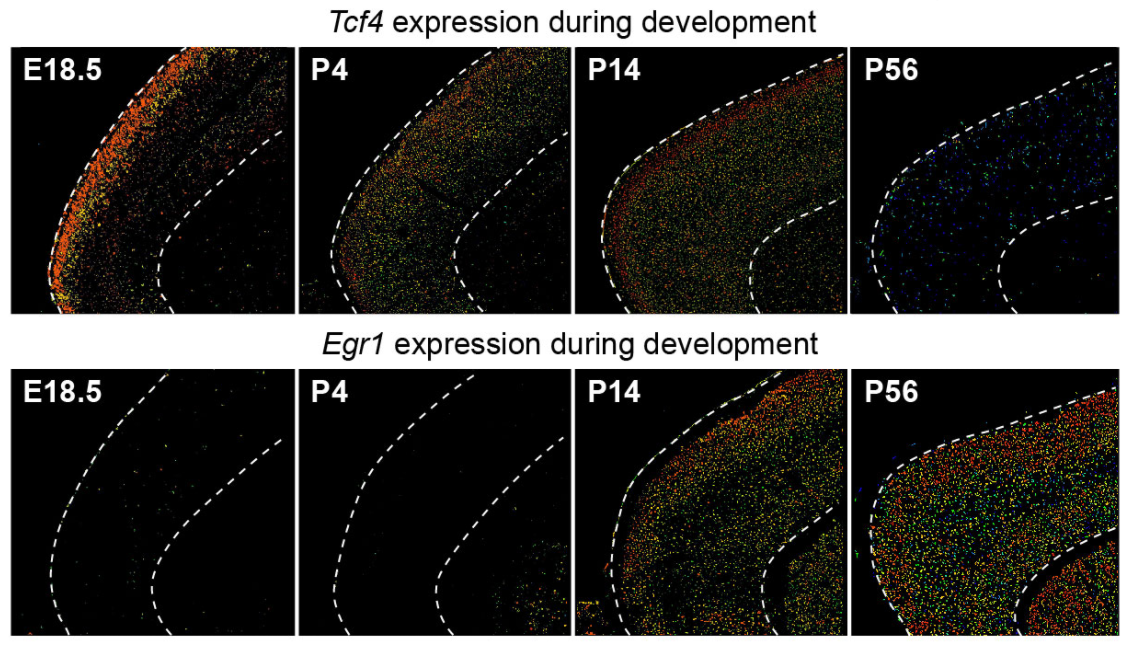
## Figure_6F
```{r}
knitr::include_graphics("./images/Figure_6F.png", dpi = 300)
```