# Install packages
if (!requireNamespace("ggplot2", quietly = TRUE)) {
install.packages("ggplot2")
}
if (!requireNamespace("stringr", quietly = TRUE)) {
install.packages("stringr")
}
# Load packages
library(ggplot2)
library(stringr)Barplot Color Group
Hiplot website
This page is the tutorial for source code version of the Hiplot Barplot Color Group plugin. You can also use the Hiplot website to achieve no code ploting. For more information please see the following link:
The color group barplot can be used to display data values in groups, and to label different colors in sequence.
Setup
System Requirements: Cross-platform (Linux/MacOS/Windows)
Programming language: R
Dependent packages:
ggplot2;stringr
Data Preparation
Data table (three columns):
Term | Entry name, such as GO/KEGG channel name
Count | The numerical size of the entry, such as the number of genes enriched in a pathway
Type | Category to which this channel belongs: such as BP/MF/CC/KEGG
# Load data
data <- read.delim("files/Hiplot/004-barplot-color-group-data.txt", header = T)
# convert data structure
colnames(data) <- c("term", "count", "type")
data[,"term"] <- str_to_sentence(str_remove(data[,"term"], pattern = "\\w+:\\d+\\W"))
data[,"term"] <- factor(data[,"term"],
levels = data[,"term"][length(data[,"term"]):1])
data[,"type"] <- factor(data[,"type"],
levels = data[!duplicated(data[,"type"]), "type"])
# View data
data term count type
1 Immune response 20 BP
2 Defense response to bacterium 11 BP
3 Cell chemotaxis 8 BP
4 Cell adhesion 17 BP
5 Complement activation 8 BP
6 Cytokine-mediated signaling pathway 8 MF
7 Phagocytosis, engulfment 5 MF
8 Negative regulation of jak-stat cascade 5 MF
9 Epoxygenase p450 pathway 4 MF
10 Chemokine-mediated signaling pathway 6 MF
11 Negative regulation of leukocyte apoptotic process 3 MF
12 B cell receptor signaling pathway 5 MF
13 Cellular response to tumor necrosis factor 6 CC
14 Positive regulation of chemotaxis 3 CC
15 Positive regulation of angiogenesis 6 CC
16 Collagen fibril organization 4 CC
17 Positive regulation of homotypic cell-cell adhesion 20 KEGG
18 Regulation of cell projection assembly 14 KEGG
19 Prostate epithelial cord elongation 10 KEGG
20 Bile acid catabolic process 8 KEGG
21 Cellular response to drug 7 KEGG
22 Glycosaminoglycan metabolic process 4 KEGGVisualization
# Barplot Color Group
p <- ggplot(data = data, aes(x = term, y = count, fill = type)) +
geom_bar(stat = "identity", width = 0.8) +
theme_bw() +
xlab("Count") +
ylab("Term") +
guides(fill = guide_legend(title="Type")) +
ggtitle("Barplot Color Group") +
coord_flip() +
theme_classic() +
scale_fill_manual(values = c("#E64B35FF","#4DBBD5FF","#00A087FF","#3C5488FF")) +
theme(text = element_text(family = "Arial"),
plot.title = element_text(size = 12,hjust = 0.5),
axis.title = element_text(size = 12),
axis.text = element_text(size = 10),
axis.text.x = element_text(angle = 0, hjust = 0.5,vjust = 1),
legend.position = "right",
legend.direction = "vertical",
legend.title = element_text(size = 10),
legend.text = element_text(size = 10))
p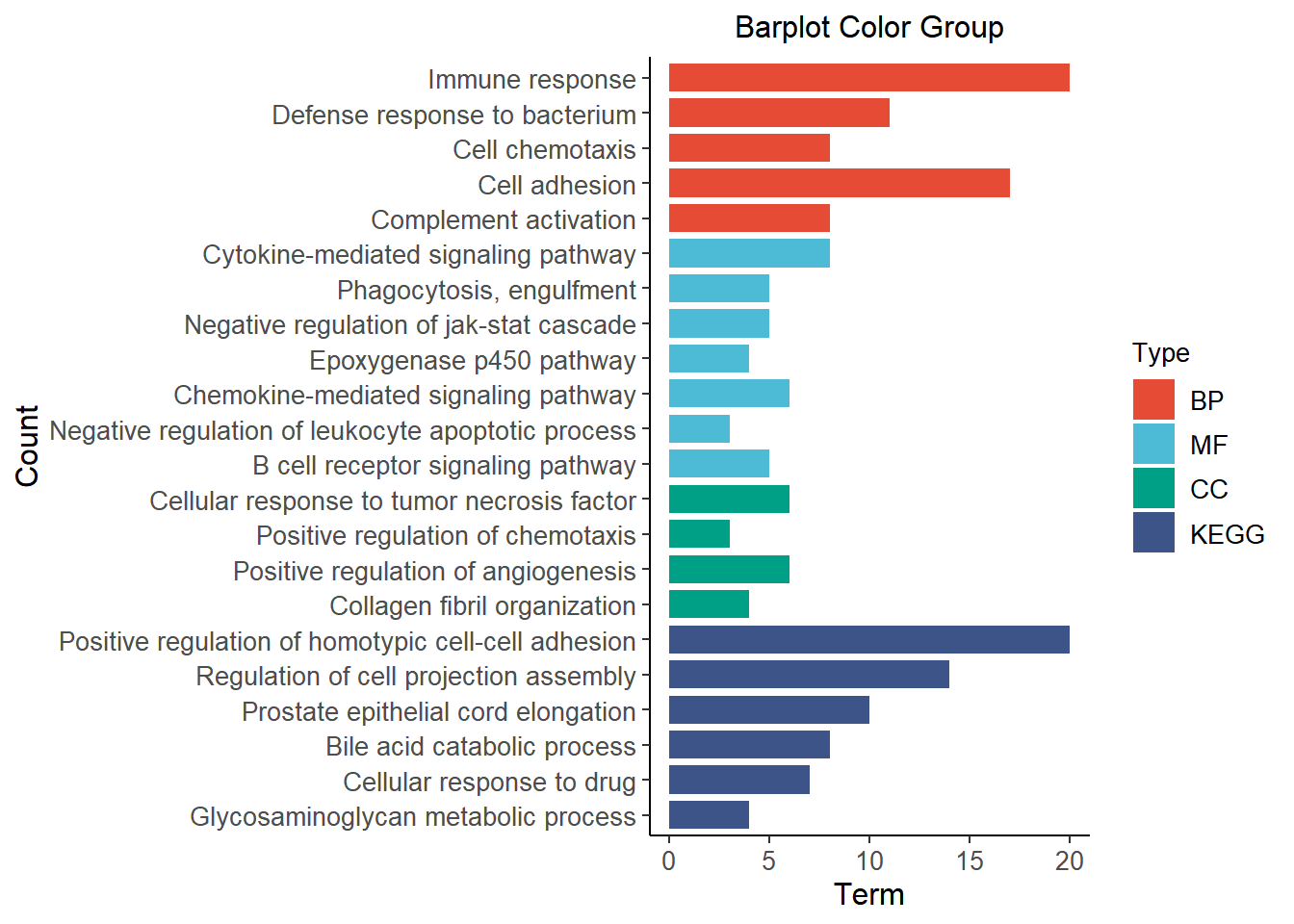
This plot visualizes the results of GO/KEGG pathway enrichment analysis.
