# Install packages
if (!requireNamespace("ggrepel", quietly = TRUE)) {
install.packages("ggrepel")
}
if (!requireNamespace("ggplot2", quietly = TRUE)) {
install.packages("ggplot2")
}
if (!requireNamespace("RColorBrewer", quietly = TRUE)) {
install.packages("RColorBrewer")
}
# Load packages
library(ggrepel)
library(ggplot2)
library(RColorBrewer)Gene Ranking Dotplot
Note
Hiplot website
This page is the tutorial for source code version of the Hiplot Gene Ranking Dotplot plugin. You can also use the Hiplot website to achieve no code ploting. For more information please see the following link:
Gene expression ranking visualization.
Setup
System Requirements: Cross-platform (Linux/MacOS/Windows)
Programming language: R
Dependent packages:
ggrepel;ggplot2;RColorBrewer
Data Preparation
# Load data
data <- read.delim("files/Hiplot/061-gene-rank-data.txt", header = T)
# Convert data structure
## ordered by log2FoldChange and pvalue
data <- data[order(-data$log2FC, data$pvalue), ]
## add the rank column
data$rank <- 1:nrow(data)
## get the top n up and down gene for labeling
top_n <- 5
top_n_up <- rownames(head(data, top_n))
top_n_down <- rownames(tail(data, top_n))
genes_to_label <- c(top_n_up, top_n_down)
data2 <- data[genes_to_label, ]
# View data
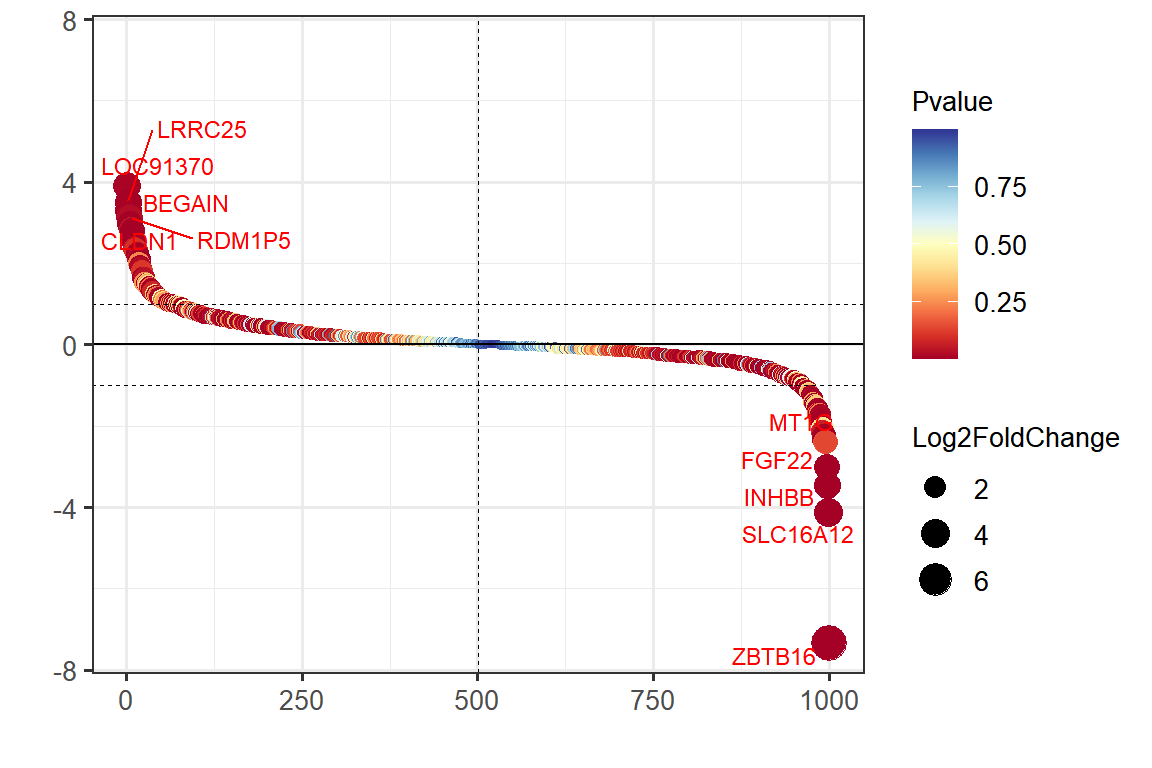
head(data) gene log2FC pvalue rank
514 LOC91370 3.889820 8.478310e-03 1
293 LRRC25 3.492413 5.000000e-05 2
470 BEGAIN 3.312996 3.347634e-03 3
159 RDM1P5 3.125070 1.538852e-02 4
194 CLDN1 3.096516 2.622163e-02 5
626 KCNS1 2.949729 2.770000e-09 6Visualization
# Gene Ranking Dotplot
p <-
ggplot(data, aes(rank, log2FC, color = pvalue, size = abs(log2FC))) +
geom_point() +
scale_color_gradientn(colours = colorRampPalette(brewer.pal(11,'RdYlBu'))(100)) +
geom_hline(yintercept = c(-1, 1), linetype = 2, size = 0.3) +
geom_hline(yintercept = 0, linetype = 1, size = 0.5) +
geom_vline(xintercept = median(data$rank), linetype = 2, size = 0.3) +
geom_text_repel(data = data2, aes(rank, log2FC, label = gene),
size = 3, color = "red") +
xlab("") + ylab("") +
ylim(c(-max(abs(data$log2FC)), max(abs(data$log2FC)))) +
labs(color = "Pvalue", size = "Log2FoldChange") +
theme_bw(base_size = 12) +
theme(text = element_text(family = "Arial"),
plot.title = element_text(size = 12,hjust = 0.5),
axis.title = element_text(size = 12),
axis.text = element_text(size = 10),
axis.text.x = element_text(angle = 0, hjust = 0.5,vjust = 1),
legend.position = "right",
legend.direction = "vertical",
legend.title = element_text(size = 10),
legend.text = element_text(size = 10))
p