# Install packages
if (!requireNamespace("ComplexHeatmap", quietly = TRUE)) {
install.packages("ComplexHeatmap")
}
# Load packages
library(ComplexHeatmap)Corrplot Big Data
Note
Hiplot website
This page is the tutorial for source code version of the Hiplot Corrplot Big Data plugin. You can also use the Hiplot website to achieve no code ploting. For more information please see the following link:
The correlation heat map is a graph that analyzes the correlation between two or more variables.
Setup
System Requirements: Cross-platform (Linux/MacOS/Windows)
Programming language: R
Dependent packages:
ComplexHeatmap
Data Preparation
The loaded data are the gene names and the expression of each sample.
# Load data
data <- read.table("files/Hiplot/013-big-corrplot-data.txt", header = T)
# convert data structure
data <- data[!is.na(data[, 1]), ]
idx <- duplicated(data[, 1])
data[idx, 1] <- paste0(data[idx, 1], "--dup-", cumsum(idx)[idx])
rownames(data) <- data[, 1]
data <- data[, -1]
str2num_df <- function(x) {
x[] <- lapply(x, function(l) as.numeric(l))
x
}
tmp <- t(str2num_df(data))
corr <- round(cor(tmp, use = "na.or.complete", method = "pearson"), 3)
# View data
head(corr) RGL4 MPP7 UGCG CYSTM1 ANXA2 ENDOD1 ARHGAP24 CST7 HIST1H2BM
RGL4 1.000 0.914 0.929 0.936 -0.592 -0.908 0.888 0.949 -0.603
MPP7 0.914 1.000 0.852 0.907 -0.543 -0.862 0.762 0.899 -0.656
UGCG 0.929 0.852 1.000 0.956 -0.440 -0.791 0.854 0.840 -0.694
CYSTM1 0.936 0.907 0.956 1.000 -0.358 -0.762 0.812 0.852 -0.632
ANXA2 -0.592 -0.543 -0.440 -0.358 1.000 0.826 -0.660 -0.723 0.541
ENDOD1 -0.908 -0.862 -0.791 -0.762 0.826 1.000 -0.907 -0.961 0.709
EREG EMP1 NFAM1 SLC40A1 CD52 HIST1H2BH PFKFB3 SNORD116-20 STX11
RGL4 -0.021 -0.495 0.859 0.506 -0.704 -0.680 0.889 0.188 0.953
MPP7 -0.196 -0.447 0.898 0.648 -0.734 -0.770 0.842 0.048 0.915
UGCG 0.153 -0.358 0.858 0.361 -0.671 -0.711 0.943 0.202 0.951
CYSTM1 0.074 -0.272 0.866 0.339 -0.612 -0.683 0.933 0.225 0.985
ANXA2 0.222 0.902 -0.662 -0.668 0.775 0.626 -0.463 0.375 -0.374
ENDOD1 0.191 0.713 -0.872 -0.611 0.854 0.791 -0.814 0.141 -0.787
SYNE2 TCN1
RGL4 0.780 0.889
MPP7 0.795 0.888
UGCG 0.922 0.927
CYSTM1 0.908 0.973
ANXA2 -0.327 -0.249
ENDOD1 -0.657 -0.708Visualization
# Corrplot Big Data
p <- ComplexHeatmap::Heatmap(
corr, col = colorRampPalette(c("#4477AA","#FFFFFF","#BB4444"))(50),
clustering_distance_rows = "euclidean",
clustering_method_rows = "ward.D2",
clustering_distance_columns = "euclidean",
clustering_method_columns = "ward.D2",
show_column_dend = FALSE, show_row_dend = FALSE,
column_names_gp = gpar(fontsize = 8),
row_names_gp = gpar(fontsize = 8)
)
p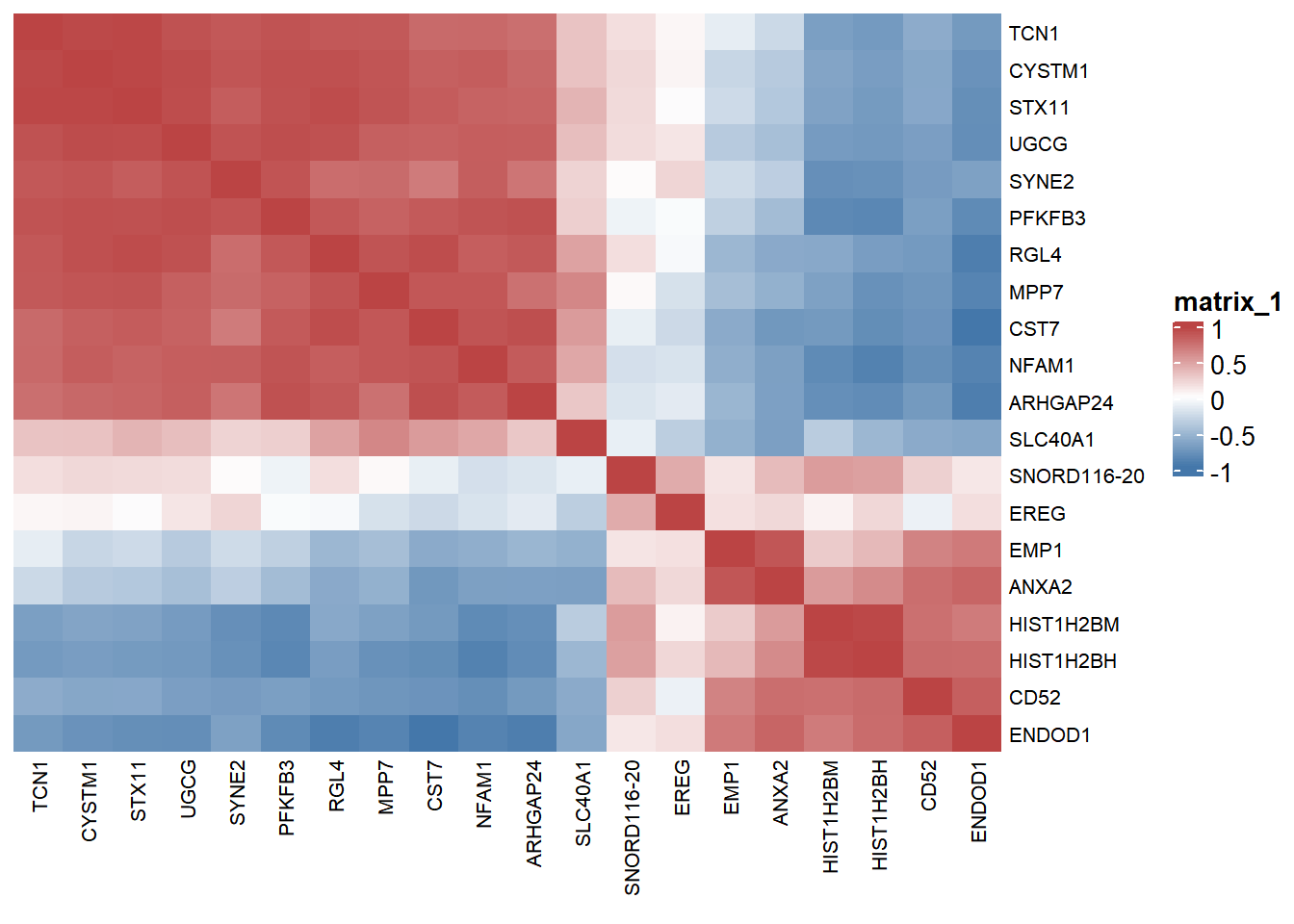
Red indicates positive correlation between two genes, blue indicates negative correlation between two genes, and the number in each cell indicates correlation coefficient.
