# Install packages
if (!requireNamespace("Mfuzz", quietly = TRUE)) {
install_github("MatthiasFutschik/Mfuzz")
}
if (!requireNamespace("ggplotify", quietly = TRUE)) {
install.packages("ggplotify")
}
if (!requireNamespace("RColorBrewer", quietly = TRUE)) {
install.packages("RColorBrewer")
}
# Load packages
library(Mfuzz)
library(ggplotify)
library(RColorBrewer)Gene Cluster Trend
Hiplot website
This page is the tutorial for source code version of the Hiplot Gene Cluster Trend plugin. You can also use the Hiplot website to achieve no code ploting. For more information please see the following link:
The gene cluster trend is used to display different gene expression trend with multiple lines showing the similar expression patterns in each cluster.
Setup
System Requirements: Cross-platform (Linux/MacOS/Windows)
Programming language: R
Dependent packages:
Mfuzz;ggplotify;RColorBrewer
Data Preparation
The loaded data are a gene expression matrix with each row represent a gene and each column represent a time-point sample.
# Load data
data <- read.delim("files/Hiplot/062-gene-trend-data.txt", header = T)
# Convert data structure
## Convert a gene expression matrix to an ExpressionSet object
row.names(data) <- data[,1]
data <- data[,-1]
data <- as.matrix(data)
eset <- new("ExpressionSet", exprs = data)
## Filter genes with more than 25% missing values
eset <- filter.NA(eset, thres=0.25)0 genes excluded.## Remove genes with small differences between samples based on standard deviation
eset <- filter.std(eset, min.std=0, visu = F)0 genes excluded.## Data Standardization
eset <- standardise(eset)
## Set the number of clusters
c <- 6
## Evaluate the optimal m value
m <- mestimate(eset)
## Perform mfuzz clustering
cl <- mfuzz(eset, c = c, m = m)
# View data
head(data) Time1 Time2 Time3
Gene1 0.1774993 1.6563226 -1.15259948
Gene2 -0.5037254 -0.5207024 0.46416071
Gene3 0.1050310 0.6079246 0.72893247
Gene4 -1.1791537 0.4340085 0.41061745
Gene5 0.8368975 -0.7047414 -1.46114720
Gene6 0.2611762 0.1351524 -0.01890809Visualization
# Gene Cluster Trend
p <- as.ggplot(function(){
mfuzz.plot2(
eset,
cl,
xlab = "Time",
ylab = "Expression changes",
mfrow = c(2,(c/2+0.5)),
colo = "fancy",
centre = T,
centre.col = "red",
time.labels = colnames(eset),
x11=F)
})
p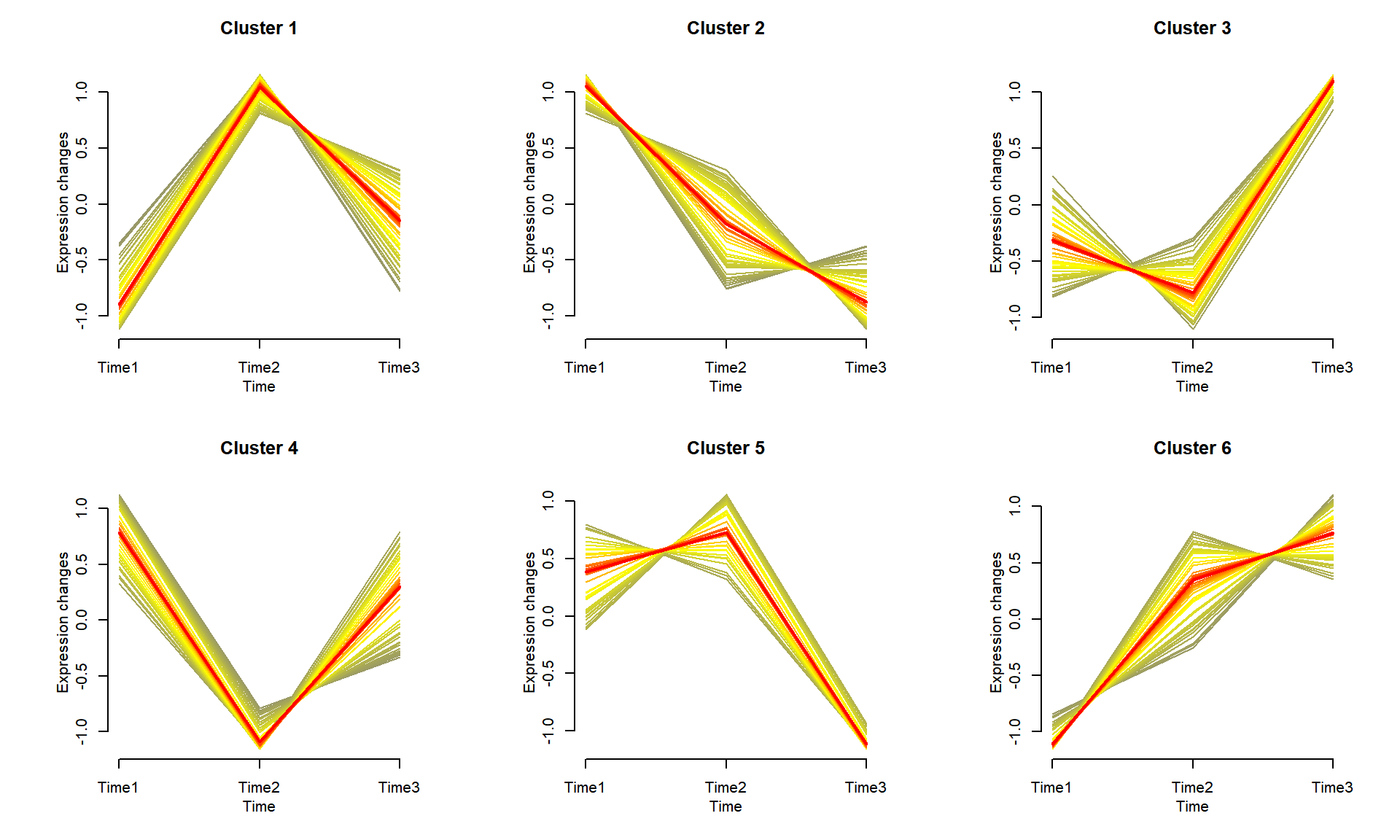
As shown in the example figure, the genes are clustered into different groups, with each group showing similar expression patterns across different time-points. The average expression trend is highlighted in each cluster.
