# Install packages
if (!requireNamespace("clusterProfiler", quietly = TRUE)) {
install_github("YuLab-SMU/clusterProfiler")
}
# Load packages
library(clusterProfiler)DIY GSEA
Note
Hiplot website
This page is the tutorial for source code version of the Hiplot DIY GSEA plugin. You can also use the Hiplot website to achieve no code ploting. For more information please see the following link:
Make your geneset.
Setup
System Requirements: Cross-platform (Linux/MacOS/Windows)
Programming language: R
Dependent packages:
clusterProfiler
Data Preparation
# Load data
data1 <- read.delim("files/Hiplot/044-diy-gsea-data1.txt", header = T)
data2 <- read.delim("files/Hiplot/044-diy-gsea-data2.txt", header = T)
# convert data structure
data1[,2] <- as.numeric(data1[,2])
geneList <- data1[,2]
names(geneList) <- data1[,1]
geneList <- sort(geneList, decreasing = TRUE)
term <- data.frame(term=data2[,1], gene=data2[,2])
# View data
head(term) term gene
1 GO_ADAPTIVE_IMMUNE_RESPONSE ADAM17
2 GO_ADAPTIVE_IMMUNE_RESPONSE AICDA
3 GO_ADAPTIVE_IMMUNE_RESPONSE ALCAM
4 GO_ADAPTIVE_IMMUNE_RESPONSE ANXA1
5 GO_ADAPTIVE_IMMUNE_RESPONSE BATF
6 GO_ADAPTIVE_IMMUNE_RESPONSE BCL10Visualization
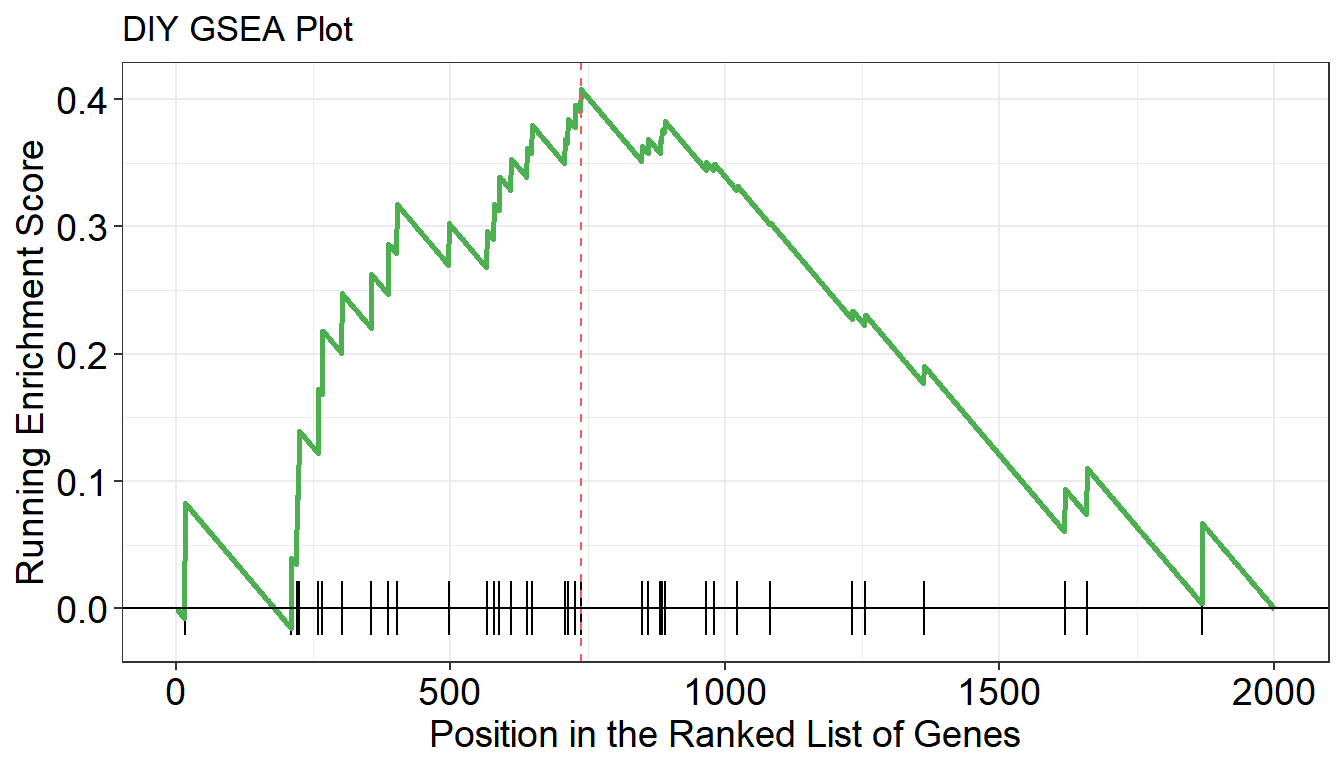
# DIY GSEA
y <- clusterProfiler::GSEA(geneList, TERM2GENE = term, pvalueCutoff = 1)
p <- gseaplot(
y,
y@result$Description[1],
color = "#000000",
by = "runningScore",
color.line = "#4CAF50",
color.vline= "#FA5860",
title = "DIY GSEA Plot",
)
p