# 安装包
if (!requireNamespace("ggplot2", quietly = TRUE)) {
install.packages("ggplot2")
}
if (!requireNamespace("dplyr", quietly = TRUE)) {
install.packages("dplyr")
}
if (!requireNamespace("ggpubr", quietly = TRUE)) {
install.packages("ggpubr")
}
if (!requireNamespace("ggthemes", quietly = TRUE)) {
install.packages("ggthemes")
}
# 加载包
library(ggplot2)
library(dplyr)
library(ggpubr)
library(ggthemes)半小提琴图
注记
Hiplot 网站
本页面为 Hiplot Half Violin 插件的源码版本教程,您也可以使用 Hiplot 网站实现无代码绘图,更多信息请查看以下链接:
半小提琴图是在保留小提琴图右半部分图形的基础上,将左侧部分换成数据频次计数图形,也是用于显示数据分布及概率密度的统计图表。
环境配置
系统: Cross-platform (Linux/MacOS/Windows)
编程语言: R
依赖包:
ggplot2;dplyr;ggpubr;ggthemes
数据准备
载入数据为载入数据为数据集 (不同肿瘤中基因名称及表达水平)。
# 加载数据
data <- read.delim("files/Hiplot/085-half-violin-data.txt", header = T)
# 整理数据格式
colnames(data) <- c("Value", "Group")
data[, 2] <- factor(data[, 2], levels = unique(data[, 2]))
# 查看数据
head(data) Value Group
1 12.10228 AML
2 12.61382 AML
3 12.52741 AML
4 12.67990 AML
5 12.64837 AML
6 12.12146 AML可视化
# 半小提琴图
geom_flat_violin <- function(
mapping = NULL, data = NULL, stat = "ydensity", position = "dodge",
trim = TRUE, scale = "area", show.legend = NA, inherit.aes = TRUE, ...) {
ggplot2::layer(data = data, mapping = mapping, stat = stat,
geom = geom_flat_violin_proto, position = position,
show.legend = show.legend, inherit.aes = inherit.aes,
params = list(trim = trim, scale = scale, ...))
}
"%||%" <- function(a, b) {
if (!is.null(a)) {
a
} else {
b
}
}
geom_flat_violin_proto <-
ggproto("geom_flat_violin_proto", Geom,
setup_data = function(data, params) {
data$width <- data$width %||%
params$width %||% (resolution(data$x, FALSE) * 0.9)
data %>%
dplyr::group_by(.data = ., group) %>%
dplyr::mutate(.data = ., ymin = min(y), ymax = max(y), xmin = x,
xmax = x + width / 2)
},
draw_group = function(data, panel_scales, coord) {
data <- base::transform(data, xminv = x,
xmaxv = x + violinwidth * (xmax - x))
newdata <- base::rbind(
dplyr::arrange(.data = base::transform(data, x = xminv), y),
dplyr::arrange(.data = base::transform(data, x = xmaxv), -y))
newdata <- rbind(newdata, newdata[1, ])
ggplot2:::ggname("geom_flat_violin",
GeomPolygon$draw_panel(newdata, panel_scales, coord))
},
draw_key = draw_key_polygon,
default_aes = ggplot2::aes(weight = 1, colour = "grey20", fill = "white",
size = 0.5, alpha = NA, linetype = "solid"),
required_aes = c("x", "y")
)
p <- ggplot(data = data, aes(Group, Value, fill = Group)) +
geom_flat_violin(alpha = 1, scale = "count", trim = FALSE) +
geom_boxplot(width = 0.05, fill = "white", alpha = 1,
outlier.colour = NA, position = position_nudge(0.05)) +
stat_summary(fun = mean, geom = "point", fill = "white", shape = 21, size = 2,
position = position_nudge(0.05)) +
geom_dotplot(alpha = 1, binaxis = "y", dotsize = 0.5, stackdir = "down",
binwidth = 0.1, position = position_nudge(-0.025)) +
theme(legend.position = "none") +
xlab(colnames(data)[2]) +
ylab(colnames(data)[1]) +
guides(fill = F) +
ggtitle("Half Violin Plot") +
scale_fill_manual(values = c("#e04d39","#5bbad6","#1e9f86")) +
theme_stata() +
theme(text = element_text(family = "Arial"),
plot.title = element_text(size = 12,hjust = 0.5),
axis.title = element_text(size = 12),
axis.text = element_text(size = 10),
axis.text.x = element_text(angle = 0, hjust = 0.5,vjust = 1),
legend.position = "right",
legend.direction = "vertical",
legend.title = element_text(size = 10),
legend.text = element_text(size = 10))
p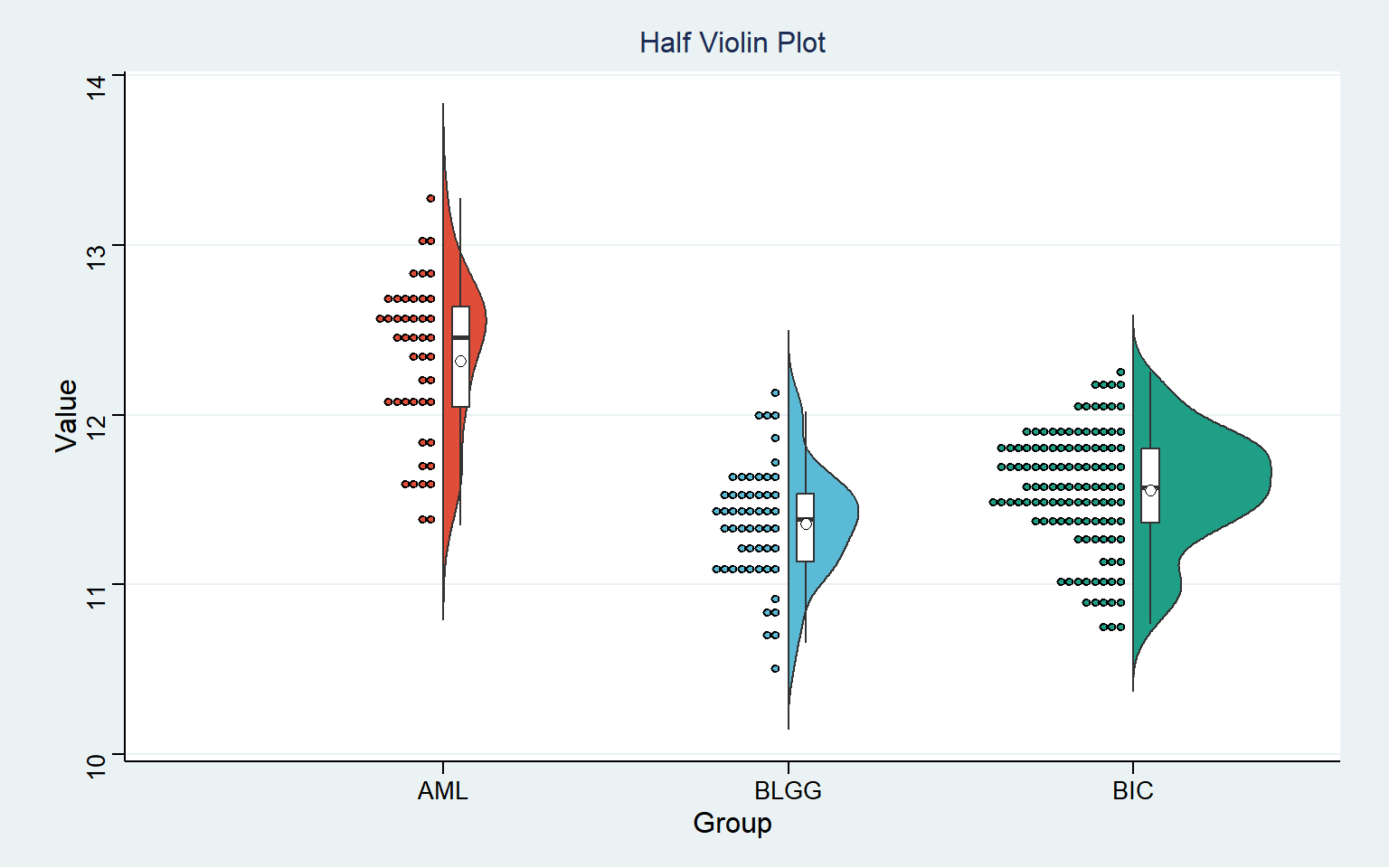
半小提琴图可以反映数据分布,同箱形图类似,方框中黑色横线显示各肿瘤中基因表达水平的中位数, 白色方框中上下框边代表数据集中的上,下四分位点;左半面可观测数值点的分布状况;小提琴图还可以反映数据密度,数据集数据越集中则图形越胖。图示中 BLGG 组中的基因表达分布更集中,BIC 组次之,AML 组则分布最分散。