# 安装包
if (!requireNamespace("limma", quietly = TRUE)) {
install.packages("limma")
}
if (!requireNamespace("Seurat", quietly = TRUE)) {
install.packages("Seurat")
}
if (!requireNamespace("ggplot2", quietly = TRUE)) {
install.packages("ggplot2")
}
# 加载包
library(limma)
library(Seurat)
library(ggplot2)堆叠小提琴图
注记
Hiplot 网站
本页面为 Hiplot Stack Violin 插件的源码版本教程,您也可以使用 Hiplot 网站实现无代码绘图,更多信息请查看以下链接:
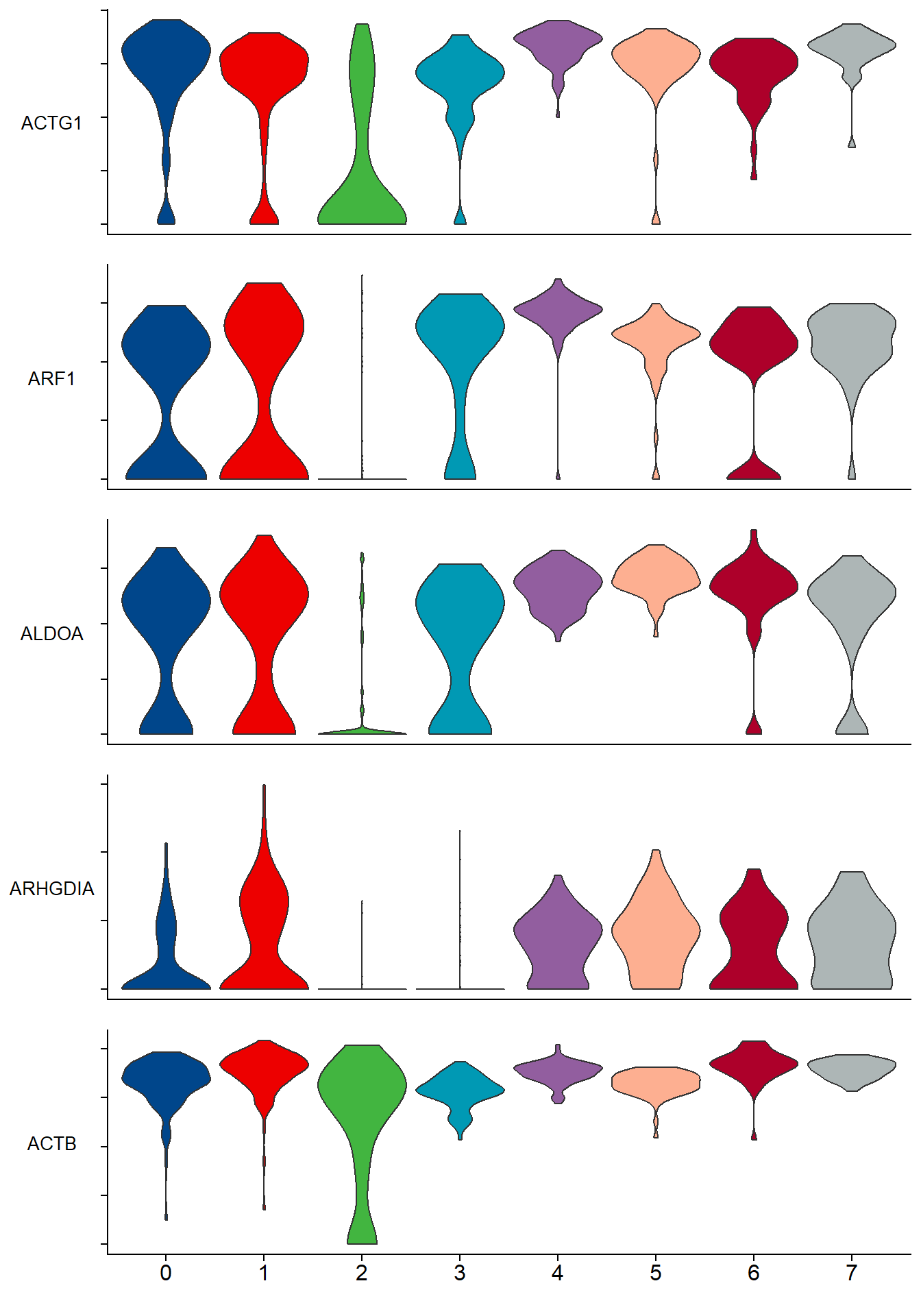
单细胞转录组(single cell RNA-Seq)分析中每个簇关键基因的表达。
环境配置
系统: Cross-platform (Linux/MacOS/Windows)
编程语言: R
依赖包:
limma;Seurat;ggplot2
数据准备
基因表达矩阵。单细胞转录组分析中所有细胞和组的基因表达矩阵(单细胞RNA序列)。
# 加载数据
data <- read.delim("files/Hiplot/166-stack-violin-data.txt", header = T)
# 整理数据格式
data <- as.matrix(data)
rownames(data) <- data[, 1]
exp <- data[, 2:ncol(data)]
dimnames <- list(
rownames(exp),
colnames(exp)
)
data <- matrix(as.numeric(as.matrix(exp)),
nrow = nrow(exp),
dimnames = dimnames
)
data <- avereps(data,
ID = rownames(data)
)
## 将矩阵转换为Seurat对象,并对数据进行过滤
pbmc <- CreateSeuratObject(
counts = data,
project = "seurat",
min.cells = 0,
min.features = 0,
names.delim = "_",
)
## 使用PercentageFeatureSet函数计算线粒体基因的百分比
pbmc[["percent.mt"]] <- PercentageFeatureSet(
object = pbmc,
pattern = "^MT-"
)
## 对数据进行过滤
pbmc <- subset(
x = pbmc,
subset = nFeature_RNA > 50 & percent.mt < 5
)
## 对数据进行标准化
pbmc <- NormalizeData(
object = pbmc,
normalization.method = "LogNormalize",
scale.factor = 10000, verbose = F
)
## 提取那些在细胞间变异系数较大的基因
pbmc <- FindVariableFeatures(
object = pbmc,
selection.method = "vst",
nfeatures = 1500, verbose = F
)
## PCA降维之前的标准预处理步骤
pbmc <- ScaleData(pbmc)
pbmc <- RunPCA(
object = pbmc,
npcs = 20,
pc.genes = VariableFeatures(object = pbmc)
)
## 每个PC的p值分布和均匀分布
pbmc <- JackStraw(
object = pbmc,
num.replicate = 100
)
pbmc <- ScoreJackStraw(
object = pbmc,
dims = 1:20
)
## 计算邻接距离
pbmc <- FindNeighbors(
object = pbmc,
dims = 1:20
)
## 对细胞分组,优化标准模块化
pbmc <- FindClusters(
object = pbmc,
resolution = 0.5
)Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 653
Number of edges: 18908
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8684
Number of communities: 8
Elapsed time: 0 seconds## TSNE聚类
pbmc <- RunTSNE(
object = pbmc,
dims = 1:20
)
## 寻找差异表达的特征
log_fc_filter <- 0.5
adj_pval_filter <- 0.05
pbmc_markers <- FindAllMarkers(
object = pbmc,
only.pos = FALSE,
min.pct = 0.25,
logfc.threshold = log_fc_filter
)
pbmc_sig_markers <- pbmc_markers[(abs(as.numeric(
as.vector(pbmc_markers$avg_logFC)
)) > log_fc_filter &
as.numeric(as.vector(pbmc_markers$p_val_adj)) <
adj_pval_filter), ]
# 查看数据
head(data[,1:5]) PT089_P1_A01 PT089_P1_A02 PT089_P1_A03 PT089_P1_A04 PT089_P1_A05
A1BG 0.00 0 0 0 0
A1BG-AS1 0.00 0 0 0 0
A1CF 0.00 0 0 1 0
A2M 0.00 0 0 339 0
A2M-AS1 0.00 0 0 0 0
A2ML1 1.08 0 0 0 0可视化
# 堆叠小提琴图
## 定义绘图函数
modify_vlnplot <- function(obj,
feature,
pt.size = 0,
plot.margin = unit(c(-0.75, 0, -0.75, 0), "cm"),
...) {
p <- VlnPlot(obj,
features = feature,
pt.size = pt.size,
...
)
p <- p +
xlab("") +
ylab(feature) +
theme(
axis.text.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_line(),
axis.title.y = element_text(angle = 0, vjust = 0.5),
plot.margin = plot.margin,
text = element_text(
family = "Arial"
),
plot.title = element_blank(),
axis.title = element_text(
size = 10
),
legend.position = "none",
legend.direction = "vertical",
legend.title = element_text(
size = 10
),
legend.text = element_text(
size = 10
)
) +
scale_fill_manual(values = c("#00468BFF","#ED0000FF","#42B540FF","#0099B4FF",
"#925E9FFF","#FDAF91FF","#AD002AFF","#ADB6B6FF"))
return(p)
}
## main function
stacked_vln_plot <- function(obj,
features,
pt.size = 0,
plot.margin = unit(c(-0.75, 0, -0.75, 0), "cm"),
...) {
plot_list <- purrr::map(
features,
function(x) {
modify_vlnplot(
obj = obj,
feature = x,
...
)
}
)
plot_list[[length(plot_list)]] <- plot_list[[length(plot_list)]] +
theme(
axis.text.x = element_text(),
axis.ticks.x = element_line()
)
p <- patchwork::wrap_plots(
plotlist = plot_list,
ncol = 1
)
return(p)
}
## 绘图
p <- stacked_vln_plot(pbmc, c("ACTG1","ARF1","ALDOA","ARHGDIA","ACTB"), pt.size = 0)
p