# 安装包
if (!requireNamespace("tidyverse", quietly = TRUE)) {
install.packages("tidyverse")
}
if (!requireNamespace("readxl", quietly = TRUE)) {
install.packages("readxl")
}
if (!requireNamespace("ggrepel", quietly = TRUE)) {
install.packages("ggrepel")
}
# 加载包
library(tidyverse)
library(readxl)
library(ggrepel)火山图
火山图用于两组之间的比较,得到两组之间的上调/下调,筛选依据为 p 值和 FC 值,转换为 -logP 值和 log2(FC) 值。导入数据可以是微生物组的 OTU 表格或 ASV 表格,或者转录组基因表达的表格,或者是代谢组学的 features 表格等多组学数据。
示例
环境配置
系统要求: 跨平台(Linux/MacOS/Windows)
编程语言:R
依赖包:
tidyverse;readxl;ggrepel
数据准备
我们导入来自 omicshare 的火山图示例数据。
# 读取Excel数据
data <- read_excel("files/volcano.eg.xlsx")
# 重命名列名(处理特殊字符)
data <- data %>%
rename(log2FC = "log2 Ratio(WT0/LOG)", Pvalue = "Pvalue")
# 处理 p-value 为 0 的情况(避免计算-Inf)
data <- data %>%
mutate(log10P = -log10(Pvalue + 1e-300)) # 确保处理了 P=0 的情况
# 转为数值型,并处理转换失败的值(如无效字符)
data <- data %>%
mutate(
log2FC = as.numeric(log2FC) # 转换失败的值会变为 NA
)
# 查找导致转换失败的原始值
data %>%
filter(is.na(log2FC)) %>%
select(log2FC) # 查看这些行的 log2FC 原始值# A tibble: 0 × 1
# ℹ 1 variable: log2FC <dbl># 根据实际情况修复数据(例如替换或删除异常值)
# 示例:将"Inf"替换为实际数值或过滤掉
data <- data %>%
mutate(
log2FC = ifelse(log2FC == "Inf", 100, log2FC), # 根据需求调整
log2FC = as.numeric(log2FC)
) %>%
filter(!is.na(log2FC)) # 删除无法修复的行
# 定义显著性(同时满足P值<0.05且|log2FC|>1)
# 定义显著性分类(上调、下调、无显著)
data <- data %>%
mutate(
significant = case_when(
Pvalue < 0.05 & log2FC > 2 ~ "Upregulated", # 上调(红色)
Pvalue < 0.05 & log2FC < -2 ~ "Downregulated", # 下调(绿色)
TRUE ~ "Not significant" # 无显著(灰色)
)
)
# 查看数据结构
head(data, 5)# A tibble: 5 × 10
GeneID LOG_count WT0_count LOG_rpkm WT0_rpkm log2FC Pvalue FDR
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Unigene0000003 17243 17525 932. 942. 0.0168 5.15e-1 6.73e-1
2 Unigene0000004 65 101 4.12 6.37 0.629 6.64e-3 1.79e-2
3 Unigene0000005 909 984 36.9 39.7 0.108 1.26e-1 2.25e-1
4 Unigene0000006 1376 1082 74.7 58.5 -0.353 1.17e-8 6.86e-8
5 Unigene0000007 121 73 15.7 9.42 -0.736 7.42e-4 2.45e-3
# ℹ 2 more variables: significant <chr>, log10P <dbl>可视化
1. 基础火山图
# 基础火山图
p <-
ggplot(data, aes(x = log2FC, y = -log10(Pvalue))) + # 初步绘制火山图
# 绘制散点,按显著分类着色
geom_point(aes(color = significant), alpha = 0.6, size = 1.5) +
# 设置颜色映射(上调红,下调绿,无显著灰)
scale_color_manual(
values = c("Upregulated" = "red", "Downregulated" = "green", "Not significant" = "gray"),
name = "Significance" # 图例标题
) +
# 添加筛选阈值线
geom_vline(xintercept = c(-2, 2), linetype = "dashed", color = "green", linewidth = 0.5) + # log2FC阈值线
geom_hline(yintercept = -log10(0.05), linetype = "dashed", color = "blue", linewidth = 0.5) + # p值阈值线
# 调整坐标轴和标题
labs(x = "log2(Fold Change)", y = "-log10(P-value)",
title = "Volcano Plot with Thresholds") +
theme_minimal() +
theme(plot.title = element_text(hjust = 0.5, face = "bold"), # 标题居中加粗
legend.position = "right") # 图例位置
p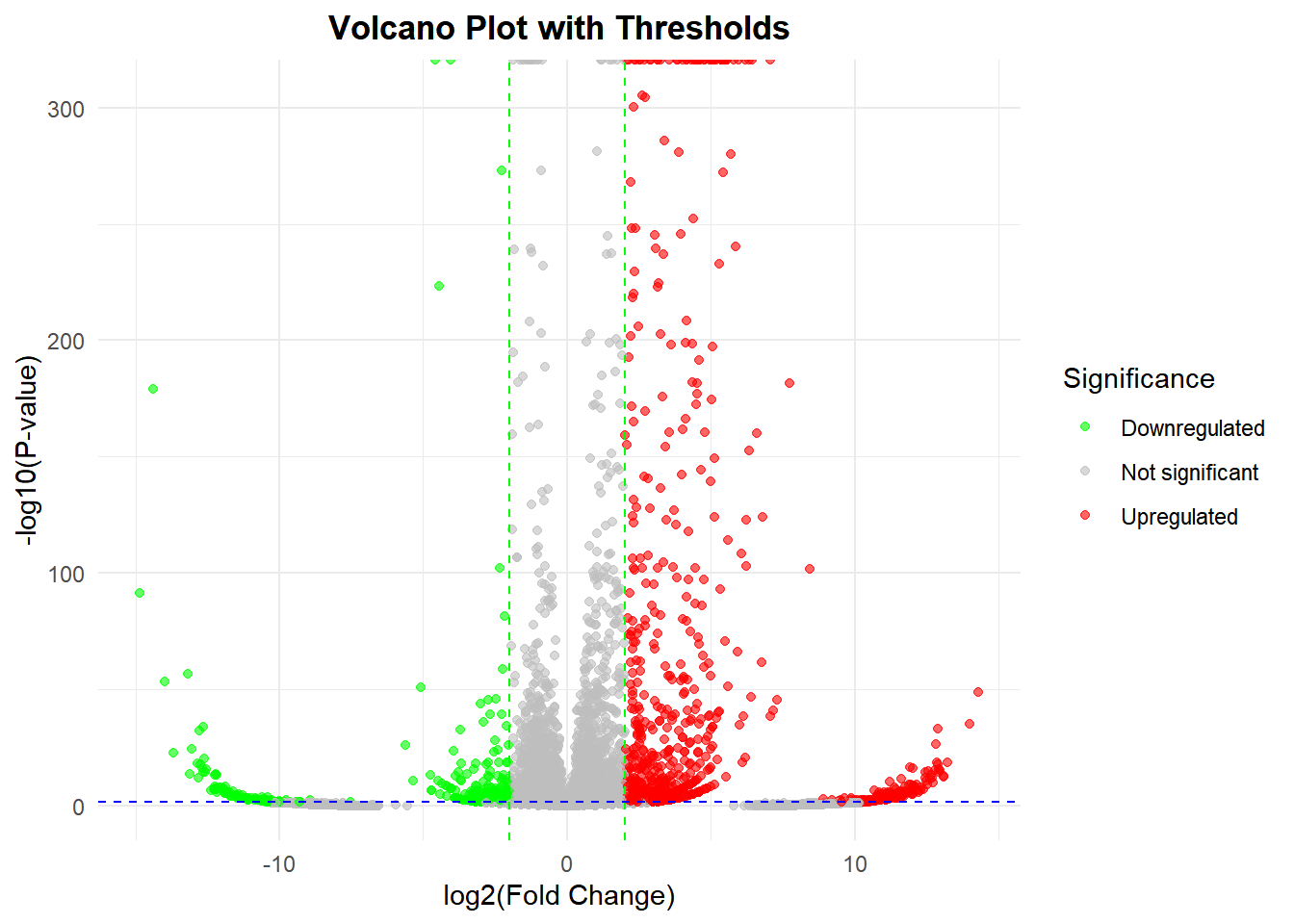
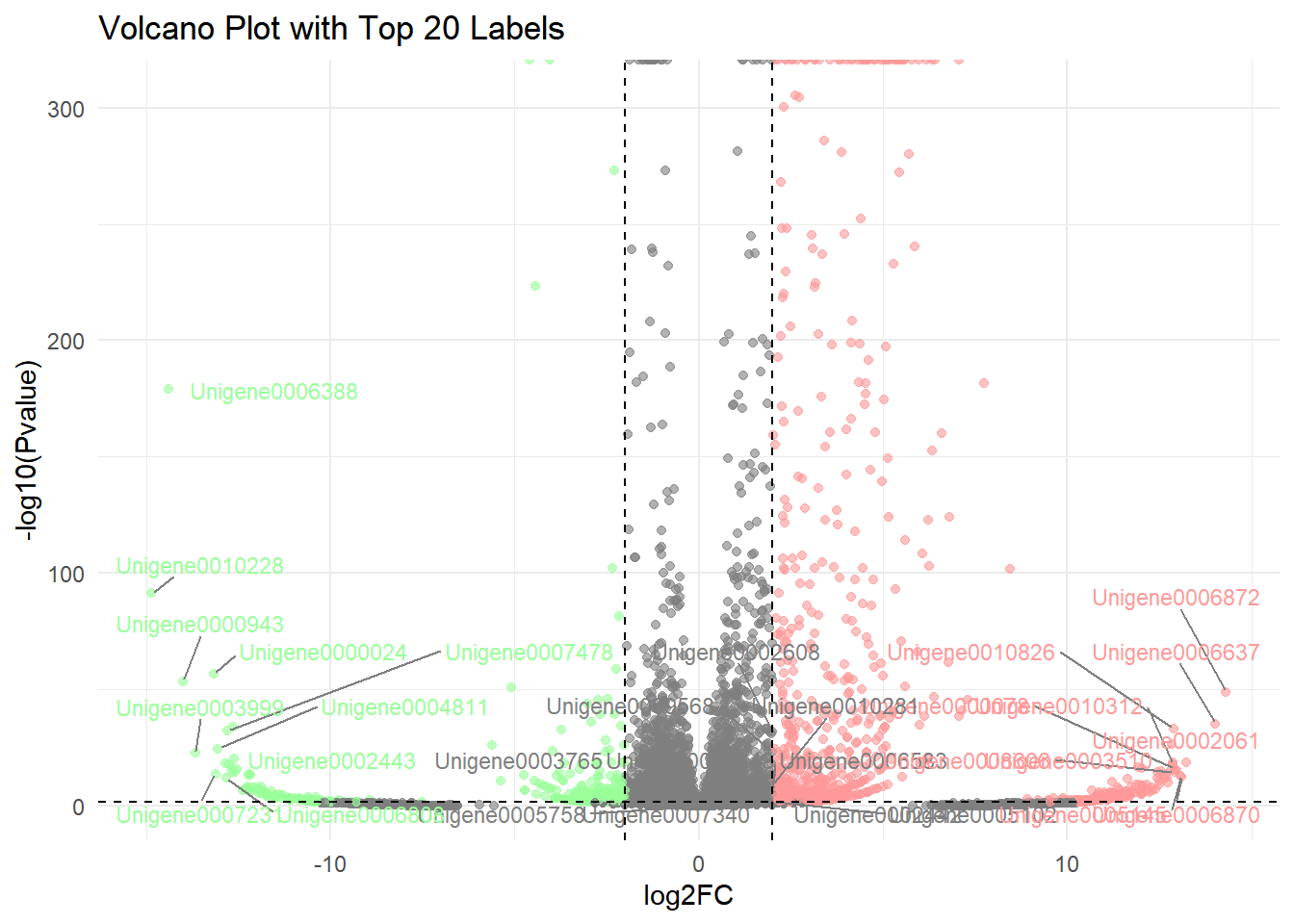
2. 带标签火山图
# 添加标签
# 步骤1:筛选需标注的基因
label_data <- data %>%
filter(Pvalue < 0.05) %>% # 筛选显著基因
group_by(significant) %>% # 按上下调控分组
top_n(10, abs(log2FC)) %>% # 取 log2FC 绝对值最大的 10 个
ungroup()
# 步骤2:定义标签颜色(浅红和浅绿)
label_colors <- c(
"Upregulated" = "#FF9999", # 浅红色
"Downregulated" = "#99FF99" # 浅绿色
)
# 步骤3:绘制带标签的火山图
p <-
ggplot(data, aes(x = log2FC, y = -log10(Pvalue))) +
geom_point(aes(color = significant), alpha = 0.6, size = 1.5) +
# 添加基因标签(仅标注目标基因)
geom_text_repel(
data = label_data,
aes(label = GeneID, color = significant), # 假设基因名列名为 OTU ID
size = 3,
box.padding = 0.5, # 标签间距
max.overlaps = 50, # 允许最大重叠
segment.color = "grey50", # 连接线颜色
show.legend = FALSE
) +
# 设置颜色映射(原颜色+标签颜色)
scale_color_manual(
values = c("Upregulated" = "red", "Downregulated" = "green", "Not significant" = "gray"),
guide = guide_legend(override.aes = list(
color = c("red", "green", "gray"), # 图例颜色保持原色
label = "" # 图例中不显示文字
))) +
# 控制标签颜色(浅红和浅绿)
scale_color_manual(
values = label_colors,
guide = "none" # 隐藏额外图例
) +
# 保留原有阈值线和标题
geom_vline(xintercept = c(-2, 2), linetype = "dashed", color = "black") +
geom_hline(yintercept = -log10(0.05), linetype = "dashed", color = "black") +
labs(title = "Volcano Plot with Top 20 Labels") +
theme_minimal()
p