# 安装包
if (!requireNamespace("ggpubr", quietly = TRUE)) {
install.packages("ggpubr")
}
# 加载包
library(ggpubr)火山图
注记
Hiplot 网站
本页面为 Hiplot Volcano 插件的源码版本教程,您也可以使用 Hiplot 网站实现无代码绘图,更多信息请查看以下链接:
火山图是一种直观展示两个样本间基因差异表达的分布图。
环境配置
系统: Cross-platform (Linux/MacOS/Windows)
编程语言: R
依赖包:
ggpubr
数据准备
载入数据为基因名称及其对应的 logFC 和 p.value。
# 加载数据
data <- read.delim("files/Hiplot/183-volcano-data.txt", header = T)
# 整理数据格式
## 对差异p(adj.P.Val一列)进行log10转换
data[, "logP"] <- -log10(as.numeric(data[, "P.Value"]))
data[, "logFC"] <- as.numeric(data[, "logFC"])
## 新加一列Group
data[, "Group"] <- "not-significant"
## Up and down
data$Group[which((data[, "P.Value"] < 0.05) & (data$logFC >= 2))] <- "Up-regulated"
data$Group[which((data[, "P.Value"] < 0.05) & (data$logFC <= 2 * -1))] <- "Down-regulated"
## 新加一列Label
data[["Label"]] <- ""
## 对差异表达基因的p值进行从小到大排序
data <- data[order(data[, "P.Value"]), ]
## 高表达的基因中,选择adj.P.Val最小的10个
up_genes <- head(data[, "Symbol"][which(data$Group == "Up-regulated")], 10)
down_genes <- head(data[, "Symbol"][which(data$Group == "Down-regulated")], 10)
not_sig_genes <- NA
## 将up_genes和down_genes合并,并加入到Label中
deg_top_genes <- c(as.character(up_genes), as.character(not_sig_genes),
as.character(down_genes))
deg_top_genes <- deg_top_genes[!is.na(deg_top_genes)]
data$Label[match(deg_top_genes, data[, "Symbol"])] <- deg_top_genes
# 查看数据
head(data) Symbol logFC P.Value logP Group Label
1 LTB 2.580831 1.17e-14 13.93181 Up-regulated LTB
2 CDCA5 -2.326302 2.46e-13 12.60906 Down-regulated CDCA5
3 C10orf54 3.307901 3.53e-13 12.45223 Up-regulated C10orf54
4 CAPN7 2.514235 1.04e-12 11.98297 Up-regulated CAPN7
5 OIP5 -2.166620 1.43e-12 11.84466 Down-regulated OIP5
7 PKIG -1.560504 1.58e-12 11.80134 not-significant 可视化
# 火山图
options(ggrepel.max.overlaps = 100)
p <- ggscatter(data, x = "logFC", y = "logP", color = "Group",
palette = c("#2f5688", "#BBBBBB", "#CC0000"), size = 1,
alpha = 0.5, font.label = 8, repel = TRUE, label=data$Label,
xlab = "log2(Fold Change)", ylab = "-log10(P Value)",
show.legend.text = FALSE) +
ggtitle("Volcano Plot") +
geom_hline(yintercept = -log(0.05, 10), linetype = "dashed") +
geom_vline(xintercept = c(2, -2), linetype = "dashed") +
theme_bw() +
theme(text = element_text(family = "Arial"),
plot.title = element_text(size = 12,hjust = 0.5),
axis.title = element_text(size = 12),
axis.text = element_text(size = 10),
axis.text.x = element_text(angle = 0, hjust = 0.5,vjust = 1),
legend.position = "right",
legend.direction = "vertical",
legend.title = element_text(size = 10),
legend.text = element_text(size = 10))
p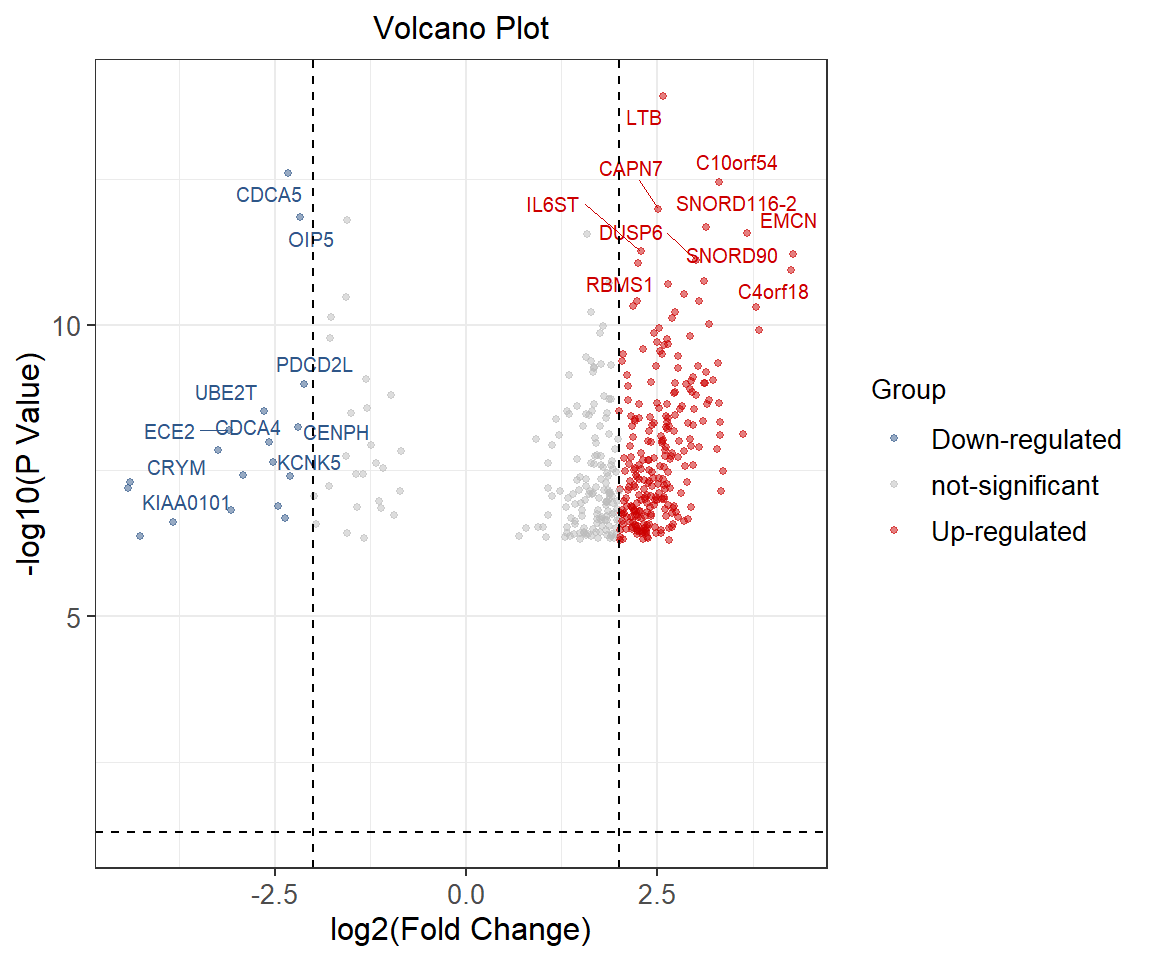
横轴用 log2(fold change)表示,差异越大的基因分布在图片两端。纵坐标用 -log10(p.value)表示,取值为 T 检验显著性 P 值的负对数。蓝色的点代表下调的基因,红色的点代表上调的基因,灰色的点代表差异不显著的基因。