# Install packages
if (!requireNamespace("ggpubr", quietly = TRUE)) {
install.packages("ggpubr")
}
# Load packages
library(ggpubr)Volcano
Hiplot website
This page is the tutorial for source code version of the Hiplot Volcano plugin. You can also use the Hiplot website to achieve no code ploting. For more information please see the following link:
The volcanogram is a visual representation of the difference in gene expression between two samples.
Setup
System Requirements: Cross-platform (Linux/MacOS/Windows)
Programming language: R
Dependent packages:
ggpubr
Data Preparation
The loaded data is the gene name and its corresponding logFC and p.value.
# Load data
data <- read.delim("files/Hiplot/183-volcano-data.txt", header = T)
# convert data structure
## Perform log10 transformation on the difference p (adj.P.Val column)
data[, "logP"] <- -log10(as.numeric(data[, "P.Value"]))
data[, "logFC"] <- as.numeric(data[, "logFC"])
## Add a new column Group
data[, "Group"] <- "not-significant"
## Up and down
data$Group[which((data[, "P.Value"] < 0.05) & (data$logFC >= 2))] <- "Up-regulated"
data$Group[which((data[, "P.Value"] < 0.05) & (data$logFC <= 2 * -1))] <- "Down-regulated"
## Add a new column Label
data[["Label"]] <- ""
## Sort the p-values of differentially expressed genes from small to large
data <- data[order(data[, "P.Value"]), ]
## Among the highly expressed genes, select the 10 with the smallest adj.P.Val
up_genes <- head(data[, "Symbol"][which(data$Group == "Up-regulated")], 10)
down_genes <- head(data[, "Symbol"][which(data$Group == "Down-regulated")], 10)
not_sig_genes <- NA
## Merge up_genes and down_genes and add them to Label
deg_top_genes <- c(as.character(up_genes), as.character(not_sig_genes),
as.character(down_genes))
deg_top_genes <- deg_top_genes[!is.na(deg_top_genes)]
data$Label[match(deg_top_genes, data[, "Symbol"])] <- deg_top_genes
# View data
head(data) Symbol logFC P.Value logP Group Label
1 LTB 2.580831 1.17e-14 13.93181 Up-regulated LTB
2 CDCA5 -2.326302 2.46e-13 12.60906 Down-regulated CDCA5
3 C10orf54 3.307901 3.53e-13 12.45223 Up-regulated C10orf54
4 CAPN7 2.514235 1.04e-12 11.98297 Up-regulated CAPN7
5 OIP5 -2.166620 1.43e-12 11.84466 Down-regulated OIP5
7 PKIG -1.560504 1.58e-12 11.80134 not-significant Visualization
# Volcano
options(ggrepel.max.overlaps = 100)
p <- ggscatter(data, x = "logFC", y = "logP", color = "Group",
palette = c("#2f5688", "#BBBBBB", "#CC0000"), size = 1,
alpha = 0.5, font.label = 8, repel = TRUE, label=data$Label,
xlab = "log2(Fold Change)", ylab = "-log10(P Value)",
show.legend.text = FALSE) +
ggtitle("Volcano Plot") +
geom_hline(yintercept = -log(0.05, 10), linetype = "dashed") +
geom_vline(xintercept = c(2, -2), linetype = "dashed") +
theme_bw() +
theme(text = element_text(family = "Arial"),
plot.title = element_text(size = 12,hjust = 0.5),
axis.title = element_text(size = 12),
axis.text = element_text(size = 10),
axis.text.x = element_text(angle = 0, hjust = 0.5,vjust = 1),
legend.position = "right",
legend.direction = "vertical",
legend.title = element_text(size = 10),
legend.text = element_text(size = 10))
p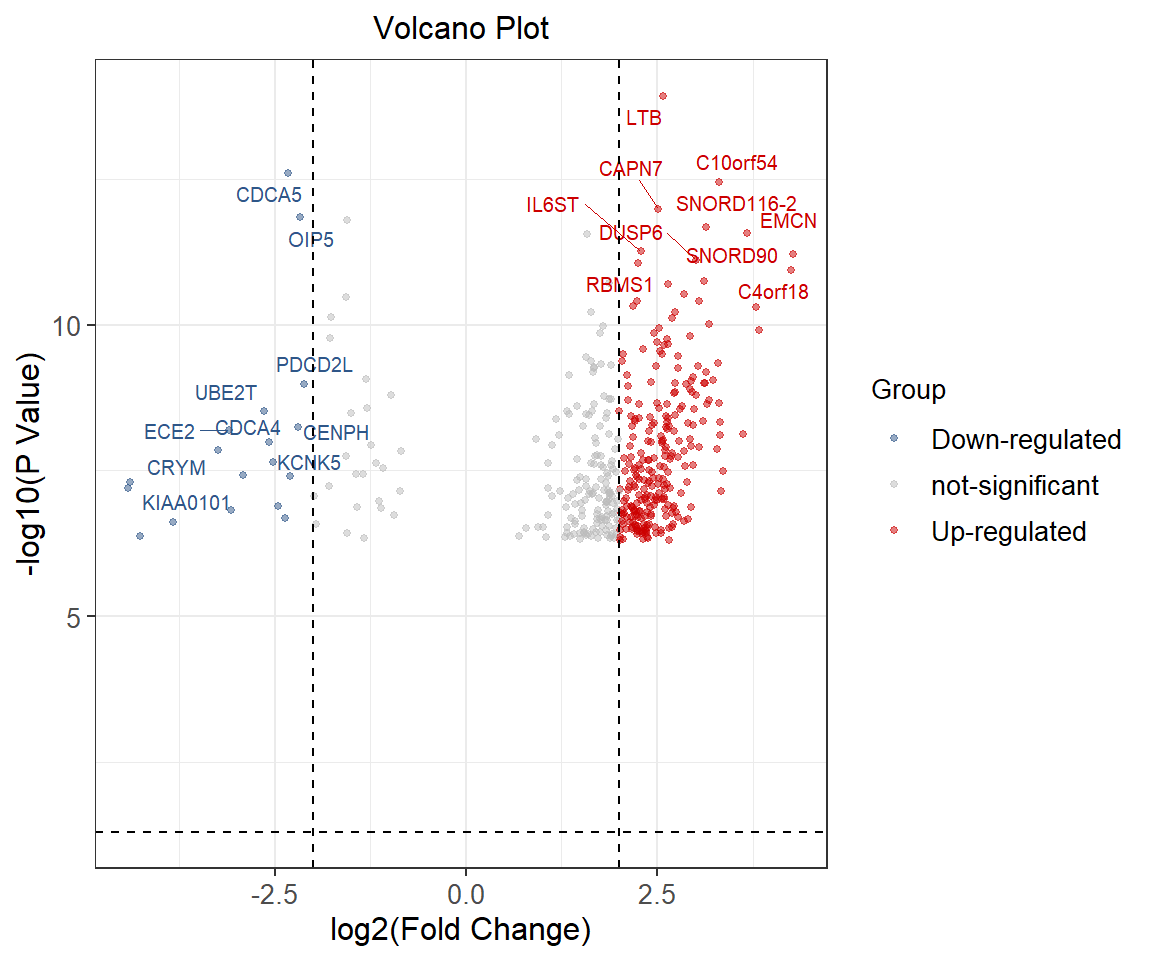
The horizontal axis is denoted by log2 (fold change), and the more different genes are distributed at both ends of the picture.The ordinate is denoted by -log10 (p.value) and is the negative log of the P value of T test significance.Blue dots represent down-regulated genes, red dots represent up-regulated genes, and gray dots represent genes that are not significantly different.
